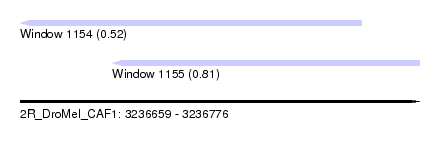
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,236,659 – 3,236,776 |
| Length | 117 |
| Max. P | 0.808498 |

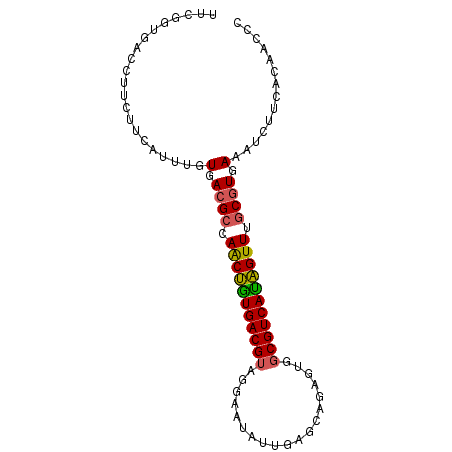
| Location | 3,236,659 – 3,236,759 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 79.03 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -18.92 |
| Energy contribution | -18.73 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3236659 100 - 20766785 UUUGUGACGCCAACUGUGACGUAGGAAUAUUGAGCUGAGUGGCGUCAUAGUUUGCGUGAAAUCUUGACAAACCCA-GCAAAUGAAUAUUUACAGCUCUGUG ..((((((((((...((...(((....)))...))....))))))))))((((((.((...............))-)))))).......((((....)))) ( -22.36) >DroSec_CAF1 5809 100 - 1 UUUGUGACGCCAACUAUGACGUAGGAAUAUUGAGCAGAGUGGCGUCAUAGUUUGCGUGAAAUCUUCACAACCCCA-GCAAAUGAAUAUUUACAUCUCUGUG ..((((((((((.((.((...(((.....)))..)).))))))))))))((((((((((.....)))).......-))))))................... ( -23.81) >DroSim_CAF1 9919 100 - 1 UUUGUGACGCCAACUGUGACGUAGGAAUAUUGAGCAGAGUGGCGUCAUAGUUUGCGUGAAAUCUUCACAACCCUA-GCAAAUGAAUAUUUACAGCUCUGUG ..((((((((((.((((...(((....)))...))))..))))))))))((((((((((.....)))).......-)))))).......((((....)))) ( -27.61) >DroEre_CAF1 5545 99 - 1 -UUGUGACGCCAACUGUGACGUAGGAAUAUUAAGCACUGUGGCGUCAUAGUUUGCGUGAAAUCUUCACAACCGCCUG-AAAUGAAUACUUACAACUCUGUG -((((((.((.(((((((((((.((...........))...))))))))))).))((((.....)))).........-..........))))))....... ( -24.60) >DroYak_CAF1 5610 99 - 1 --UGUGACGCCAACUGUGACGUAGAAACAUUAAGCACUGUGGCGUCAUAGUUUGCGUGAAAUUUUCACAACCGCCUGCAAAUGAAUAUUUACAACUCUGUA --(((((.((.(((((((((((....(((........))).))))))))))).))((((.....))))....................)))))........ ( -25.90) >DroPer_CAF1 30781 81 - 1 UUUGUGACGACAGCAGUGACGAUGG---AUUGACCAUGUCGGCGUCACUGUUUGCAUGACAUCUCCUUCACCUUA---AGCUGAAUA-------------- .(..((.(((..(((((((((.(.(---((.......))).)))))))))))))))..).......((((.(...---.).))))..-------------- ( -20.30) >consensus UUUGUGACGCCAACUGUGACGUAGGAAUAUUGAGCAGAGUGGCGUCAUAGUUUGCGUGAAAUCUUCACAACCCCA_GCAAAUGAAUAUUUACAACUCUGUG ....(.((((.(((((((((((...................))))))))))).)))).).......................................... (-18.92 = -18.73 + -0.19)

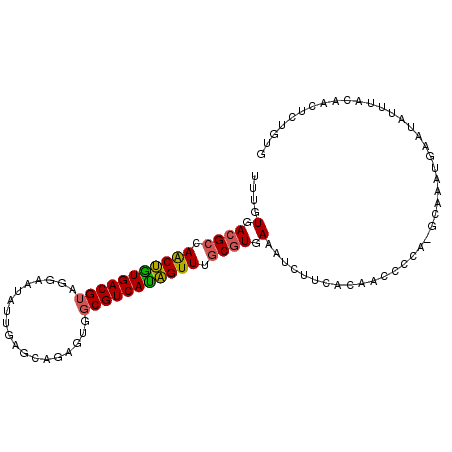
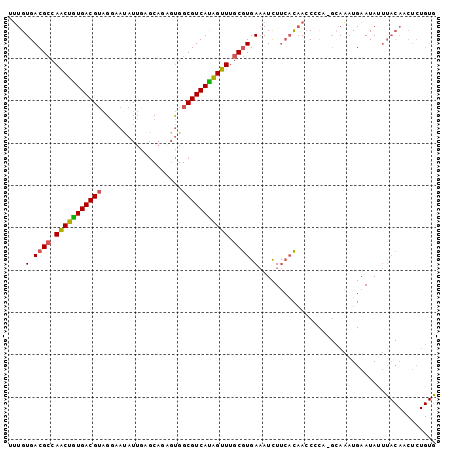
| Location | 3,236,686 – 3,236,776 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 80.71 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -18.92 |
| Energy contribution | -18.73 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3236686 90 - 20766785 CUUUGUGACCUUCUUCAUUUGUGACGCCAACUGUGACGUAGGAAUAUUGAGCUGAGUGGCGUCAUAGUUUGCGUGAAAUCUUGACAAACC ..............(((....(.((((.(((((((((((.((.........)).....))))))))))).)))).).....)))...... ( -22.50) >DroSec_CAF1 5836 90 - 1 UUCGGUGACCUUCUUCAUUUGUGACGCCAACUAUGACGUAGGAAUAUUGAGCAGAGUGGCGUCAUAGUUUGCGUGAAAUCUUCACAACCC ...(((............(((((((((((.((.((...(((.....)))..)).))))))))))))).....((((.....)))).))). ( -24.10) >DroSim_CAF1 9946 90 - 1 UUCGGUGACCUUCUUCAUUUGUGACGCCAACUGUGACGUAGGAAUAUUGAGCAGAGUGGCGUCAUAGUUUGCGUGAAAUCUUCACAACCC ...(((............(((((((((((.((((...(((....)))...))))..))))))))))).....((((.....)))).))). ( -27.30) >DroEre_CAF1 5572 88 - 1 UUUUGUGACCUUCUUC--UUGUGACGCCAACUGUGACGUAGGAAUAUUAAGCACUGUGGCGUCAUAGUUUGCGUGAAAUCUUCACAACCG ..((((((.....(((--.....((((.(((((((((((.((...........))...))))))))))).)))))))....))))))... ( -25.20) >DroYak_CAF1 5638 87 - 1 UUUUGUGACCUUUUUC---UGUGACGCCAACUGUGACGUAGAAACAUUAAGCACUGUGGCGUCAUAGUUUGCGUGAAAUUUUCACAACCG ..((((((....((((---....((((.(((((((((((....(((........))).))))))))))).))))))))...))))))... ( -29.20) >DroPer_CAF1 30792 82 - 1 CCCGGU-----UUUCCAUUUGUGACGACAGCAGUGACGAUGG---AUUGACCAUGUCGGCGUCACUGUUUGCAUGACAUCUCCUUCACCU ...(((-----(.((((((.((.((.......)).)))))))---)..))))(((((((((........))).))))))........... ( -22.70) >consensus UUCGGUGACCUUCUUCAUUUGUGACGCCAACUGUGACGUAGGAAUAUUGAGCAGAGUGGCGUCAUAGUUUGCGUGAAAUCUUCACAACCC .....................(.((((.(((((((((((...................))))))))))).)))).).............. (-18.92 = -18.73 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:42 2006