| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,335,477 – 20,335,567 |
| Length | 90 |
| Max. P | 0.990617 |

| Location | 20,335,477 – 20,335,567 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -37.47 |
| Consensus MFE | -16.74 |
| Energy contribution | -17.43 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
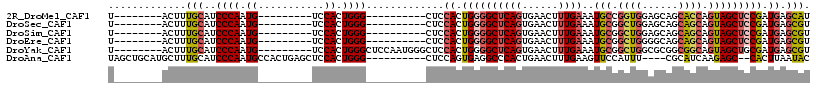
>2R_DroMel_CAF1 20335477 90 + 20766785 U--------ACUUUGCAUCCCAAUG---------UCCACUGGG----------CUCCACUGGGGCUCAGUGAACUUUGAAAUGCCGGUGGAGCAGCACCAGUAGCUCCGAUGAGCAU (--------(((..((((..(((.(---------(.(((((((----------((((...))))))))))).)).)))..)))).((((......))))))))((((....)))).. ( -37.50) >DroSec_CAF1 52052 90 + 1 U--------ACUUUGCAUCCCAAUG---------UCCACUGGG----------CUCCACUGGGGCUCAGUGAACUUUGAAAUGCGGCUGGAGCAGCAGCAGUAGCUCCGAUGAGCGU .--------....(((((..(((.(---------(.(((((((----------((((...))))))))))).)).)))..)))))((((((((.((....)).)))))....))).. ( -38.40) >DroSim_CAF1 53638 90 + 1 U--------ACUUUGCAUCCCAAUG---------UCCACUGGG----------CUCCACUGGGGCUCAGUGAACUUUGAAAUGCGGCUGGAGCAGCAGCAGUAGCUCCGAUGAGCGU .--------....(((((..(((.(---------(.(((((((----------((((...))))))))))).)).)))..)))))((((((((.((....)).)))))....))).. ( -38.40) >DroEre_CAF1 54688 90 + 1 U--------ACUUUGCAUCCCAAUG---------UCCACUGGG----------CUCCACUGGGGCUCAGUGAACUUUGAAAUGCGGCUGGGGCAGCAGCAGUAGCUCCGAUGAGCGU (--------(((.(((((..(((.(---------(.(((((((----------((((...))))))))))).)).)))..)))))((((......))))))))((((....)))).. ( -38.30) >DroYak_CAF1 53302 100 + 1 U--------ACUUUGCAUCCCAAUG---------UCCACUGGGCUCCAAUGGGCUCCACUGGGGCUCAGUGAACUUUGAAAUGCGGCUGGCGCGGCGGCAGUAGCUGCGAUGAGCGU .--------....(((((..(((.(---------(.((((((((((((.(((...))).)))))))))))).)).)))..)))))(((.(((((((.......)))))).).))).. ( -47.90) >DroAna_CAF1 51048 101 + 1 UAGCUGCAUGCUUUGCAUCCCAAUGCCACUGAGCUCCACUGGG----------CUCCAGUGAGGCCCACUGAACUUUGAAGUUCCAUUU----CGCAUCAAGAGC--CACUUAAUAC ..((((((.....)))......((((...((.((..((((((.----------..))))))..)).))..(((((....))))).....----.))))....)))--.......... ( -24.30) >consensus U________ACUUUGCAUCCCAAUG_________UCCACUGGG__________CUCCACUGGGGCUCAGUGAACUUUGAAAUGCGGCUGGAGCAGCAGCAGUAGCUCCGAUGAGCGU .............(((..((((.((...........)).)))).............((.((((((((((......))))..(((.((((......)))).))))))))).)).))). (-16.74 = -17.43 + 0.70)
| Location | 20,335,477 – 20,335,567 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -22.12 |
| Energy contribution | -22.85 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.76 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
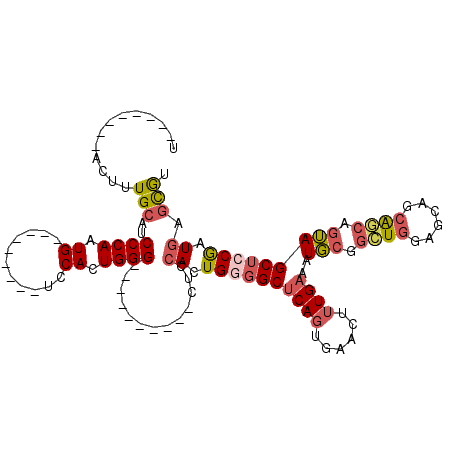
>2R_DroMel_CAF1 20335477 90 - 20766785 AUGCUCAUCGGAGCUACUGGUGCUGCUCCACCGGCAUUUCAAAGUUCACUGAGCCCCAGUGGAG----------CCCAGUGGA---------CAUUGGGAUGCAAAGU--------A ..((((....))))((((((((......)))).((((..(((.((((((((.((((....)).)----------).)))))))---------).)))..))))..)))--------) ( -38.90) >DroSec_CAF1 52052 90 - 1 ACGCUCAUCGGAGCUACUGCUGCUGCUCCAGCCGCAUUUCAAAGUUCACUGAGCCCCAGUGGAG----------CCCAGUGGA---------CAUUGGGAUGCAAAGU--------A ..((((....))))((((((((......)))).((((..(((.((((((((.((((....)).)----------).)))))))---------).)))..))))..)))--------) ( -38.60) >DroSim_CAF1 53638 90 - 1 ACGCUCAUCGGAGCUACUGCUGCUGCUCCAGCCGCAUUUCAAAGUUCACUGAGCCCCAGUGGAG----------CCCAGUGGA---------CAUUGGGAUGCAAAGU--------A ..((((....))))((((((((......)))).((((..(((.((((((((.((((....)).)----------).)))))))---------).)))..))))..)))--------) ( -38.60) >DroEre_CAF1 54688 90 - 1 ACGCUCAUCGGAGCUACUGCUGCUGCCCCAGCCGCAUUUCAAAGUUCACUGAGCCCCAGUGGAG----------CCCAGUGGA---------CAUUGGGAUGCAAAGU--------A ..((((....))))((((((((......)))).((((..(((.((((((((.((((....)).)----------).)))))))---------).)))..))))..)))--------) ( -38.60) >DroYak_CAF1 53302 100 - 1 ACGCUCAUCGCAGCUACUGCCGCCGCGCCAGCCGCAUUUCAAAGUUCACUGAGCCCCAGUGGAGCCCAUUGGAGCCCAGUGGA---------CAUUGGGAUGCAAAGU--------A ..(((...(((.((.......)).)))..))).((((..(((.((((((((.((.(((((((...))))))).)).)))))))---------).)))..)))).....--------. ( -45.30) >DroAna_CAF1 51048 101 - 1 GUAUUAAGUG--GCUCUUGAUGCG----AAAUGGAACUUCAAAGUUCAGUGGGCCUCACUGGAG----------CCCAGUGGAGCUCAGUGGCAUUGGGAUGCAAAGCAUGCAGCUA ........((--(((((..((((.----.....(((((....)))))(.(((((..((((((..----------.))))))..))))).).))))..)))((((.....)))))))) ( -38.80) >consensus ACGCUCAUCGGAGCUACUGCUGCUGCUCCAGCCGCAUUUCAAAGUUCACUGAGCCCCAGUGGAG__________CCCAGUGGA_________CAUUGGGAUGCAAAGU________A ..((((....))))....((((......)))).((((.......(((((((.....)))))))...........(((((((...........))))))))))).............. (-22.12 = -22.85 + 0.73)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:31 2006