| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,335,119 – 20,335,217 |
| Length | 98 |
| Max. P | 0.935964 |

| Location | 20,335,119 – 20,335,217 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.10 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -20.04 |
| Energy contribution | -20.68 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
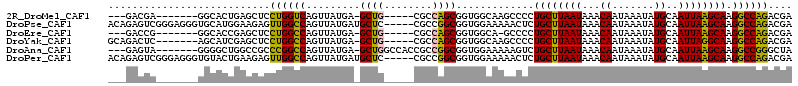
>2R_DroMel_CAF1 20335119 98 + 20766785 UCGUCUGGCCUUGCUUAAUUGCAUAUUUAUUGUUUAUUAAGCAGGGGCUUGCCACCGCUGGCG-----CAGC-UCAUAACUGACCAGGAGCUCAGUGCC-------UCGUC--- ..((..(.(((((((((((.(((.......)))..))))))))))).)..))...((..((((-----((((-((............)))))..)))))-------.))..--- ( -33.50) >DroPse_CAF1 61531 109 + 1 UCGUCUGGCCUUGCUUAAUUGCAUAUUUAUUGUUUAUUAAGCAGAGUUUUUCCACCGCCGGCG-----GAGCAUCAUAACUGGCCAACUCUUCCAUGCACCCUCCCGACUCUGU ..((.(((((.((((((((.(((.......)))..))))))))((((((..((......))..-----)))).))......))))))).......................... ( -25.20) >DroEre_CAF1 54331 97 + 1 UCGUCUGGCCUUGCUUAAUUGCAUAUUUAUUGUUUAUUAAGCAGGGGC-UGCCACCGCUGGCG-----CAGC-UCAUAACUGGCCAGGAGCUCGGUGCC-------CGGUC--- ..(.(((((((((((((((.(((.......)))..)))))))))))..-...((((((((((.-----(((.-......)))))))).....))))).)-------))).)--- ( -36.80) >DroYak_CAF1 52944 101 + 1 UCGUCUGGCCUUGCCUAAUUGCAUAUUUAUUGUUUAUUAAGCAGGGGCUUGCCACCGCUGGCG-----CAGC-UCAUAACUGGCCAGGAGCUCGAUGCU-------GAGUCUGC ...(((((((((((.((((.(((.......)))..)))).))))((((((((((....)))))-----.)))-))......))))))).(((((....)-------)))).... ( -34.90) >DroAna_CAF1 50694 103 + 1 UAGCCCGGCCUUGCUUAAUUGCAUAUUUAUUGUUUAUUAAGCAGACUUUUUCCACCGCCGGCGGUGGCCAGC-UCAUAACUGGCCGGGCGGCCAGCCCC-------UACUC--- ..((((((((.((((((((.(((.......)))..))))))))((((....((((((....))))))..)).-))......))))))))((....))..-------.....--- ( -42.70) >DroPer_CAF1 61462 109 + 1 UCGUCUGGCCUUGCUUAAUUGCAUAUUUAUUGUUUAUUAAGCAGAGUUUUUCCACCGCCGGCG-----GAGCAUCAUAACUGGCCAACUCUUCAGUACACCCUCCCGACUCUGU ..((((((((.((((((((.(((.......)))..))))))))((((((..((......))..-----)))).))......)))))(((....)))..........)))..... ( -27.10) >consensus UCGUCUGGCCUUGCUUAAUUGCAUAUUUAUUGUUUAUUAAGCAGAGGCUUGCCACCGCCGGCG_____CAGC_UCAUAACUGGCCAGGAGCUCAGUGCC_______GACUC___ ...((((((((((((((((.(((.......)))..)))))))))................((........)).........))))))).......................... (-20.04 = -20.68 + 0.64)

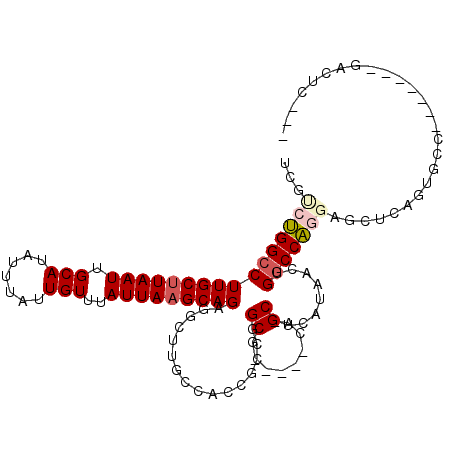
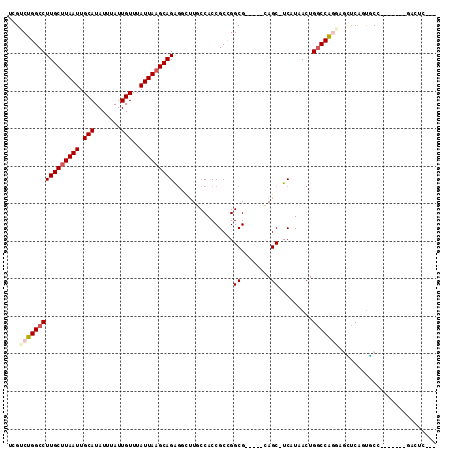
| Location | 20,335,119 – 20,335,217 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.10 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -21.64 |
| Energy contribution | -20.95 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20335119 98 - 20766785 ---GACGA-------GGCACUGAGCUCCUGGUCAGUUAUGA-GCUG-----CGCCAGCGGUGGCAAGCCCCUGCUUAAUAAACAAUAAAUAUGCAAUUAAGCAAGGCCAGACGA ---..((.-------(.(((((.....(((((((((.....-))))-----.)))))))))).)..(((..((((((((...((.......))..)))))))).)))....)). ( -28.70) >DroPse_CAF1 61531 109 - 1 ACAGAGUCGGGAGGGUGCAUGGAAGAGUUGGCCAGUUAUGAUGCUC-----CGCCGGCGGUGGAAAAACUCUGCUUAAUAAACAAUAAAUAUGCAAUUAAGCAAGGCCAGACGA .....(((.((((.((.(((((..(.(....))..))))))).)))-----)((((((((.(......).)))))................(((......))).)))..))).. ( -27.80) >DroEre_CAF1 54331 97 - 1 ---GACCG-------GGCACCGAGCUCCUGGCCAGUUAUGA-GCUG-----CGCCAGCGGUGGCA-GCCCCUGCUUAAUAAACAAUAAAUAUGCAAUUAAGCAAGGCCAGACGA ---...((-------(.(((((.....(((((((((.....-))))-----.)))))))))).).-(((..((((((((...((.......))..)))))))).)))....)). ( -33.40) >DroYak_CAF1 52944 101 - 1 GCAGACUC-------AGCAUCGAGCUCCUGGCCAGUUAUGA-GCUG-----CGCCAGCGGUGGCAAGCCCCUGCUUAAUAAACAAUAAAUAUGCAAUUAGGCAAGGCCAGACGA ...(((((-------......))).))((((((........-(((.-----.((((....)))).)))...((((((((...((.......))..)))))))).)))))).... ( -29.20) >DroAna_CAF1 50694 103 - 1 ---GAGUA-------GGGGCUGGCCGCCCGGCCAGUUAUGA-GCUGGCCACCGCCGGCGGUGGAAAAAGUCUGCUUAAUAAACAAUAAAUAUGCAAUUAAGCAAGGCCGGGCUA ---.....-------.((.((((((..(((((((((.....-)))))))((((....))))))........((((((((...((.......))..)))))))).)))))).)). ( -42.00) >DroPer_CAF1 61462 109 - 1 ACAGAGUCGGGAGGGUGUACUGAAGAGUUGGCCAGUUAUGAUGCUC-----CGCCGGCGGUGGAAAAACUCUGCUUAAUAAACAAUAAAUAUGCAAUUAAGCAAGGCCAGACGA .....(((.((((.((..((((..........))))....)).)))-----)((((((((.(......).)))))................(((......))).)))..))).. ( -26.90) >consensus ___GACUC_______GGCACUGAGCUCCUGGCCAGUUAUGA_GCUG_____CGCCAGCGGUGGAAAAACCCUGCUUAAUAAACAAUAAAUAUGCAAUUAAGCAAGGCCAGACGA ...........................((((((.........((((........)))).............((((((((...((.......))..)))))))).)))))).... (-21.64 = -20.95 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:29 2006