
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,328,332 – 20,328,437 |
| Length | 105 |
| Max. P | 0.535163 |

| Location | 20,328,332 – 20,328,437 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.76 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -18.29 |
| Energy contribution | -18.43 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20328332 105 + 20766785 -----UGCAAAACGGUGCCGUGAAUGAGUCUCGUGUUUUGAGAAAUUUAUUGAUAGUGGGCGUGGGCUUCCUAAUUGAAUUAGUGUGCCACGAAAGUUCAUGUGCUCAA------G -----.(((....((..((((((((...(((((.....))))).))))))...(((..(((....)))..))).........).)..))((....)).....)))....------. ( -25.20) >DroPse_CAF1 53326 105 + 1 -GGGGAACAAAAA---GCUGUGAAUGAGUCUCGUUUUUAGAGAAAUUUAUUGAUAGUGGGCGUGGGCUUCCUAAUUGAAUUAGUGUGCCACAAAAGUUCAUGUGCCCAG------- -.(((.(((....---(((((.(((((.((((.......))))...))))).)))))((((.(((((...(((((...)))))...))).))...)))).))).)))..------- ( -31.10) >DroGri_CAF1 58451 106 + 1 U--AAA-UAAAAAGUAGAUGUGAAUGAGUCUCGU-UUUUGAGAAAUUUAUUGAUAGUGGGCGUGGGCUUCCUAAUUGAAUUAGUGUGCCACAAAAGUUCACGCACACAC------U .--...-...........(((.(((((.(((((.-...)))))...))))).)))(((.((((((((((.(((((...)))))((.....)).)))))))))).)))..------. ( -26.20) >DroWil_CAF1 83088 106 + 1 -CUGGAAAUGA---GAAAUGUGAAUGAGUCUCGUUUUAGUGAAAAUUUAUUGAUAGUGGGCGUGGGCUUCCUAAUUGAAUUAGUGUGCCACAAAAGUUCAUACAAACAG------C -((((((((((---((.((....))...)))))))))..................((((((.(((((...(((((...)))))...))).))...)))))).....)))------. ( -23.70) >DroAna_CAF1 44244 101 + 1 --------------GAGCUGUGAAUGAGUCUCGUGUUUUGAGAAAUUUAUUGAUAGUGGGCGUGGGCUUCCUAAUUGAAUUAGUGUGCCAC-AAAGUUCAUGUGCUUGCUCAGCGG --------------..(((((.(((((.(((((.....)))))...))))).)))))((((((((.(...(((((...)))))...)))))-.((((......))))))))..... ( -27.30) >DroPer_CAF1 53192 105 + 1 -GGGGAACAAAAA---GCUGUGAAUGGGUCUCGUUUUUAGAGAAAUUUAUUGAUAGUGGGCGUGGGCUUCCUAAUUGAAUUAGUGUGCCACAAAAGUUCAUGUGCCCAG------- -.(((.(((....---(((((.(((((.((((.......))))...))))).)))))((((.(((((...(((((...)))))...))).))...)))).))).)))..------- ( -30.30) >consensus ___GGAACAAAAA_GAGCUGUGAAUGAGUCUCGUUUUUUGAGAAAUUUAUUGAUAGUGGGCGUGGGCUUCCUAAUUGAAUUAGUGUGCCACAAAAGUUCAUGUGCACAG_______ ..................(((.(((((.((((.......))))...))))).)))((((((.(((((...(((((...)))))...))).))...))))))............... (-18.29 = -18.43 + 0.14)

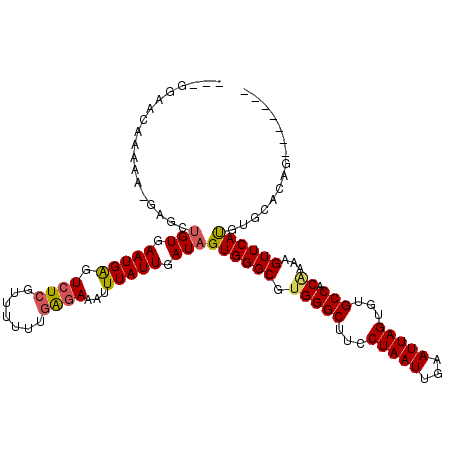
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:27 2006