
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,326,543 – 20,326,637 |
| Length | 94 |
| Max. P | 0.788379 |

| Location | 20,326,543 – 20,326,637 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.82 |
| Mean single sequence MFE | -23.37 |
| Consensus MFE | -11.61 |
| Energy contribution | -12.40 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20326543 94 - 20766785 GCCAUUGGGCCAACUCACUCGGAGACUGCCACACAUUCGCCCAUA-------------ACAGAAUGGAGUAAUUAAUUAAGGCUAACGAAAGCCUAAUUGCAGCAAU ((.(((((((...(((.....)))...(((...(((((.......-------------...)))))((....))......)))........))))))).))...... ( -22.20) >DroSec_CAF1 43228 94 - 1 GCCAUUGGGCCAACUCACUCGGCCACUGCCACACAUUCGCCCAUA-------------AUAGAAUGGAGUAAUUAAUUAAGGCUUACGAAAGCCUAAUUGCAGCAAU ...((((((((.........)))).((((.........((((((.-------------.....)))).))....(((((.(((((....)))))))))))))))))) ( -23.90) >DroSim_CAF1 44753 94 - 1 ACCAUUGGGCCAACUCACUCGGCCACUGCCACACAUUCGCCCAUA-------------ACAGAAUGGAGUAAUUAAUUAAGGCUAACGAAAGCCUAAUUGCAGCAAU ...((((((((.........)))).((((.........((((((.-------------.....)))).))....(((((.((((......))))))))))))))))) ( -23.20) >DroEre_CAF1 46059 94 - 1 GCCAUUGGGCCAGUUCCCUCGGCCACUGCCACACAUGCGCCCAUU-------------ACAGAAUGGAGUAAUUAAUUACGGCUAACGAAAGCCUAAUUGCAGCAAU ...((((((((((....)).)))).((((......(((..(((((-------------....))))).)))...(((((.((((......))))))))))))))))) ( -25.70) >DroYak_CAF1 43929 73 - 1 GCCAUUGGGCC---------------------ACAUUCGCCCAUA-------------ACAGAAUGGAGCAAUUAAUUACGGCUAACGAAAGCCUAAUUGCAGCAAU .(((((((((.---------------------......)))))..-------------.....)))).(((((((.....((((......)))))))))))...... ( -20.31) >DroPer_CAF1 50872 102 - 1 GGCUCCUGGCCAACG-----GUUCUCUGCCACACAUUCAUCCAUCAGGCAACUACGCCAAAGAAUGGUGUAAUUAAUUACGCCAAACGAAAGCCUAAUUACUGCAAU ((((..((((....(-----((.....)))...............((....))..)))).....((((((((....))))))))......))))............. ( -24.90) >consensus GCCAUUGGGCCAACUCACUCGGCCACUGCCACACAUUCGCCCAUA_____________ACAGAAUGGAGUAAUUAAUUAAGGCUAACGAAAGCCUAAUUGCAGCAAU .....(((((..........(((....)))........))))).........................(((((((.....((((......)))))))))))...... (-11.61 = -12.40 + 0.79)

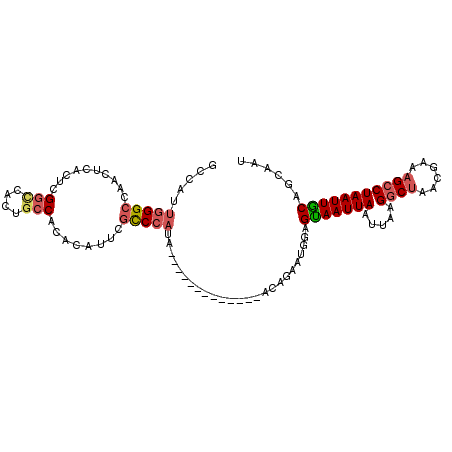
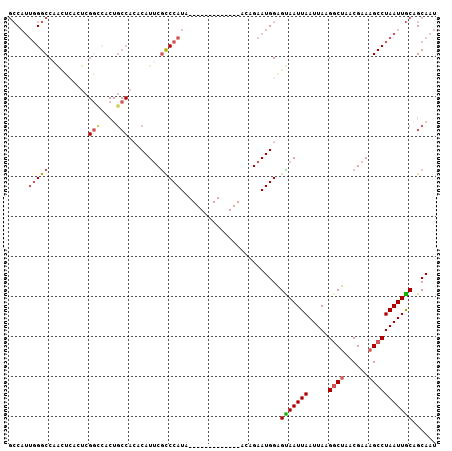
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:26 2006