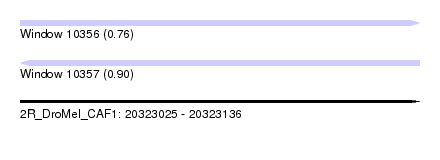
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,323,025 – 20,323,136 |
| Length | 111 |
| Max. P | 0.899838 |

| Location | 20,323,025 – 20,323,136 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.40 |
| Mean single sequence MFE | -43.52 |
| Consensus MFE | -23.96 |
| Energy contribution | -24.13 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20323025 111 + 20766785 CCCAGUCGGGCAUGGACGCCCCCGACGCCCCGCACACGCCCAAGUACAUGGACGGCGGGAACACGGCG---------GCCAGCGUCACGCCCGGCAUUAACAUCCCGGGAAAGUCCGCCU ....(((((((...((((((((((....(((((...((.(((......))).)))))))....))).)---------)...)))))..)))))))..........((((....))))... ( -44.60) >DroVir_CAF1 52485 120 + 1 CACAAUCGGGCAUGGAUGCGCCGGACGCACCGCAUACGCCCAAAUAUAUGGACGGCGGCAGCACCGUGCCCGUCAGCGCCAGCAUCACGCCCGGCGUCAAUAUACCCGGCAAAUCAGCGU ......(((((((((.(((((((...((...))...((.(((......))).)).)))).))))))))))))...((((..((.....))((((.((......)))))).......)))) ( -44.40) >DroGri_CAF1 51153 114 + 1 CACAAUCGGGCAUGGAUGCGCCGGACGCACCACAUACGCCCAAAUAUAUGGACGGUGGCAGCACCUCAGC------CGCCAGCAUCACGCCCGGCGUUCAAAUACCGGGCAAGUCCGCCU .......((((.(((.((((.....))))))).....))))........((((((((((.........))------))))........((((((..(....)..))))))..)))).... ( -45.30) >DroWil_CAF1 73151 111 + 1 CGCAAUCGGGCAUGGAUGCUCCGGAUGCCCCUCAUACGCCCAAAUAUAUGGAUGGAGGGAGUACGGCC---------AGUAGCAUAACGCCCGGUGUUAAUAUACCCGGCAAAUCGGCCU .(((.(((((((....))).)))).)))(((((......(((......)))...))))).....((((---------.((......))(((.(((((....))))).))).....)))). ( -41.30) >DroMoj_CAF1 62576 120 + 1 CACAGUCGGGCAUGGAUGCACCAGAUGCACCGCAUACGCCCAAAUAUAUGGACGGCGGUAGCACGGCAGCCGUUAGCGCCAGCAUAACACCUGGCGUUAAUAUACCUGGCAAAUCAGCAU ....((((((...((.(((....(.((((((((...((.(((......))).))))))).)))).))).))(((((((((((........)))))))))))...)))))).......... ( -45.50) >DroPer_CAF1 47595 111 + 1 CCCAGUCGGGCAUGGACGCCCCCGAUGCCCCCCACACGCCCAAAUACAUGGACGGUGGCAACACCGCG---------GCCAGCGUCACGCCCGGCGUCAACAUACCCGGAAAGUCAGCCU ....(((((((...(((((..(((...............(((......))).(((((....)))))))---------)...)))))..)))))))............((........)). ( -40.00) >consensus CACAAUCGGGCAUGGAUGCCCCGGACGCACCGCAUACGCCCAAAUAUAUGGACGGCGGCAGCACCGCG_________GCCAGCAUCACGCCCGGCGUUAAUAUACCCGGCAAAUCAGCCU ....(((((((...(((((..........((((...((.(((......))).)))))).......((..........))..)))))..)))))))..........((((....))))... (-23.96 = -24.13 + 0.17)

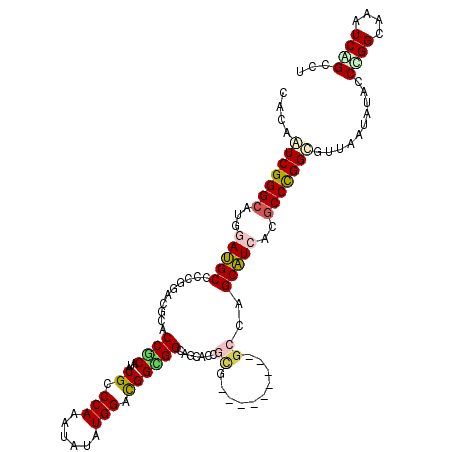
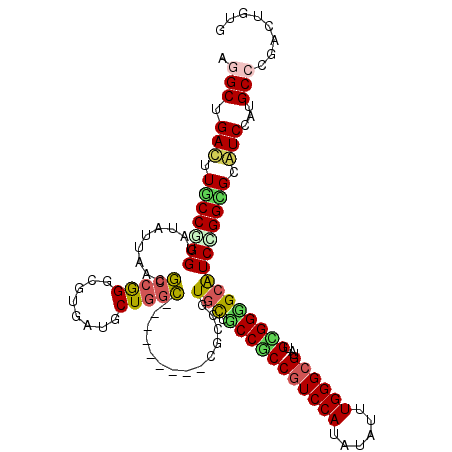
| Location | 20,323,025 – 20,323,136 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.40 |
| Mean single sequence MFE | -52.03 |
| Consensus MFE | -34.65 |
| Energy contribution | -33.98 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20323025 111 - 20766785 AGGCGGACUUUCCCGGGAUGUUAAUGCCGGGCGUGACGCUGGC---------CGCCGUGUUCCCGCCGUCCAUGUACUUGGGCGUGUGCGGGGCGUCGGGGGCGUCCAUGCCCGACUGGG ...........(((((...((....))((((((((((((...(---------(.((((((((((((((((((......)))))).))).)))))).))))))))).)))))))).))))) ( -54.80) >DroVir_CAF1 52485 120 - 1 ACGCUGAUUUGCCGGGUAUAUUGACGCCGGGCGUGAUGCUGGCGCUGACGGGCACGGUGCUGCCGCCGUCCAUAUAUUUGGGCGUAUGCGGUGCGUCCGGCGCAUCCAUGCCCGAUUGUG ..((......))(((((((..((.((((((((((......(((((((.......)))))))(((((((((((......))))))...)))))))))))))))))...)))))))...... ( -59.10) >DroGri_CAF1 51153 114 - 1 AGGCGGACUUGCCCGGUAUUUGAACGCCGGGCGUGAUGCUGGCG------GCUGAGGUGCUGCCACCGUCCAUAUAUUUGGGCGUAUGUGGUGCGUCCGGCGCAUCCAUGCCCGAUUGUG .(((((.(.((((((((........)))))))).)....(((((------((......))))))))))))((((...((((((..(((.((((((.....))))))))))))))).)))) ( -56.50) >DroWil_CAF1 73151 111 - 1 AGGCCGAUUUGCCGGGUAUAUUAACACCGGGCGUUAUGCUACU---------GGCCGUACUCCCUCCAUCCAUAUAUUUGGGCGUAUGAGGGGCAUCCGGAGCAUCCAUGCCCGAUUGCG .(((((....(((.(((........))).)))((......)))---------)))).....((((((..(((......)))..)...)))))(((..(((.(((....))))))..))). ( -41.10) >DroMoj_CAF1 62576 120 - 1 AUGCUGAUUUGCCAGGUAUAUUAACGCCAGGUGUUAUGCUGGCGCUAACGGCUGCCGUGCUACCGCCGUCCAUAUAUUUGGGCGUAUGCGGUGCAUCUGGUGCAUCCAUGCCCGACUGUG ..((......))..(((((.....((((((((((...((((((((.....)).)))).)).(((((((((((......))))))...))))))))))))))).....)))))........ ( -45.80) >DroPer_CAF1 47595 111 - 1 AGGCUGACUUUCCGGGUAUGUUGACGCCGGGCGUGACGCUGGC---------CGCGGUGUUGCCACCGUCCAUGUAUUUGGGCGUGUGGGGGGCAUCGGGGGCGUCCAUGCCCGACUGGG .......((...((((((((..((((((.((((...))))..(---------(.(((((((.((((((((((......)))))).)).)).)))))))))))))))))))))))...)). ( -54.90) >consensus AGGCUGACUUGCCGGGUAUAUUAACGCCGGGCGUGAUGCUGGC_________CGCCGUGCUGCCGCCGUCCAUAUAUUUGGGCGUAUGCGGGGCAUCCGGCGCAUCCAUGCCCGACUGUG .(((.(((.(((((((.........(((((........)))))..............(((.(((((((((((......))))))...))))))))))))))).)))...)))........ (-34.65 = -33.98 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:25 2006