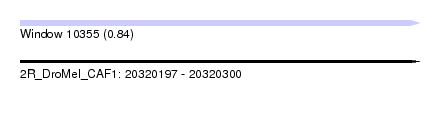
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,320,197 – 20,320,300 |
| Length | 103 |
| Max. P | 0.841263 |

| Location | 20,320,197 – 20,320,300 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 74.81 |
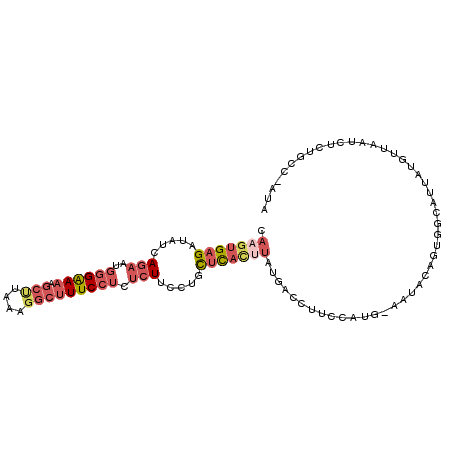
| Mean single sequence MFE | -25.11 |
| Consensus MFE | -9.99 |
| Energy contribution | -11.52 |
| Covariance contribution | 1.53 |
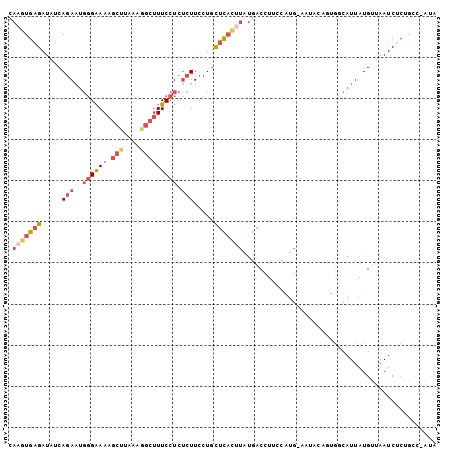
| Combinations/Pair | 1.26 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
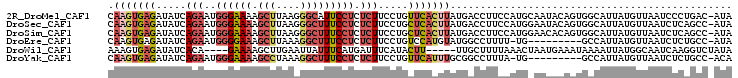
>2R_DroMel_CAF1 20320197 103 + 20766785 CAAGUGAGAUAUCAGAAUGGGAAAAGCUUAAGGGCAUUCCUCUCUUCCUGUUCACUUAUGACCUUCCAUGCAAUACAGUGGCAUUAUGUUAAUCCCUGAC-AUA .(((((((.....(((..(((((..(((....))).))))).))).....)))))))........((((........))))...(((((((.....))))-))) ( -20.90) >DroSec_CAF1 36986 103 + 1 CAAGUGAGAUAUCAGAAUGGGAAAAGCUUAAGGGCUUUCCUCUCUUCCUGCUCACUUAUGACCUUCCAUGGAAUACAGUGGCAUUAUGUUAAUCUCAGCC-AUA .(((((((.....(((..((((((.(((....))))))))).))).....)))))))....((......))......(((((...............)))-)). ( -26.16) >DroSim_CAF1 38521 103 + 1 CAAGUGAGAUAUCAGAAUGGGAAAAGCUUAAGGGCUUUCCUCUCUUCCUGCUCACUUAUGACCUUCCAUGGAACACAGUGGCAUUAUGUUAAUCUCAGCC-AUA .(((((((.....(((..((((((.(((....))))))))).))).....)))))))....((......))......(((((...............)))-)). ( -26.16) >DroEre_CAF1 39675 93 + 1 CAAGUGAGAUAUCAGAAUGGGGAAAGCUUAAAGGCUUUCCUCUCUUCCUGUCCAUGUAUGGCCUUUU-UG---------GCCAUUAUGUUAAUCUCUGCC-AUA .....(((((..(((...((((((((((....)))))))))).....)))..((((.((((((....-.)---------)))))))))...)))))....-... ( -33.30) >DroWil_CAF1 67631 95 + 1 AAAGUGAGAUAUCACA----GAAAAGCUUGAAUUAUUUCAUGAUUUCAUACUU-----UUGCUUUUAAACUAAUGAAAUAAAAUUAUGGCAAUCAAGGUCUAUA ...((((....))))(----((......((((....))))((((((((((.((-----(((.(((((......)))))))))).))))).)))))...)))... ( -15.40) >DroYak_CAF1 37482 93 + 1 CAAGUGAGAUAUCAGAAUGGGAAAAGCCUAAAGGCUUUCCUCUCUUCCUGUUCAUUUGCGGCCUUUA-UG---------GCCAUUAUGUUAAUCUCUGCC-ACA .....(((((....((((((((((((((....))))).......)))))))))......((((....-.)---------))).........)))))....-... ( -28.71) >consensus CAAGUGAGAUAUCAGAAUGGGAAAAGCUUAAAGGCUUUCCUCUCUUCCUGCUCACUUAUGACCUUCCAUG_AAUACAGUGGCAUUAUGUUAAUCUCUGCC_AUA .(((((((.....(((..((((((.(((....))))))))).))).....)))))))............................................... ( -9.99 = -11.52 + 1.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:23 2006