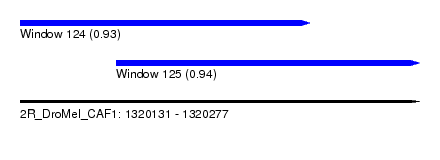
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,320,131 – 1,320,277 |
| Length | 146 |
| Max. P | 0.939216 |

| Location | 1,320,131 – 1,320,237 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.51 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -23.58 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1320131 106 + 20766785 CAAUCCACCUC----CAGCCAGGCACUCCCACC-ACAAUUCUCUAGUCCCCCCUUUC---------CACGCCUGCACCGCAAUUAUGUUUUGUGGCUUGCGGCGUGCGGACCGGAAAUUA ...(((.(((.----.....)))..........-...........((((........---------((((((.(((((((((.......))))))..))))))))).)))).)))..... ( -27.80) >DroSec_CAF1 38665 115 + 1 CCAUCC-CCCC----CAGCCACUCCCUACCACCUACAUUUCACCAGUCCCCCCAUUCCCACCAUGCCACGCCUGCACCGCAAUUAUGUUUUGUGGCUUGCGGCGUGCGGACCGGAAAUUA ......-....----...........................((.((((...(((.......))).((((((.(((((((((.......))))))..))))))))).)))).))...... ( -27.30) >DroSim_CAF1 17744 119 + 1 CAAUCC-CCCCCACCCAGCCACUCCCUACCACCUACAAUUCCACAGUCCCUCCAUUCCCACCAUGCCACGCCUGCACCGCAAUUAUGUUUUGUGGCUUGCGGCGUGCGGACCGGAAAUUA ......-...............................((((...((((...(((.......))).((((((.(((((((((.......))))))..))))))))).)))).)))).... ( -27.30) >consensus CAAUCC_CCCC____CAGCCACUCCCUACCACCUACAAUUCACCAGUCCCCCCAUUCCCACCAUGCCACGCCUGCACCGCAAUUAUGUUUUGUGGCUUGCGGCGUGCGGACCGGAAAUUA ......................................((((...((((.................((((((.(((((((((.......))))))..))))))))).)))).)))).... (-23.58 = -23.80 + 0.22)

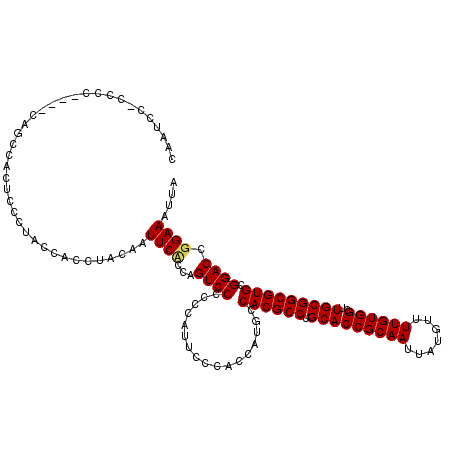
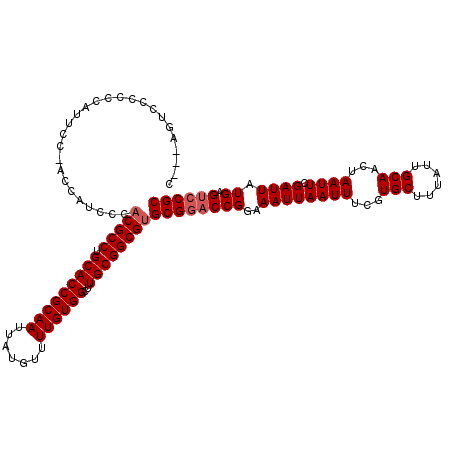
| Location | 1,320,166 – 1,320,277 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.00 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -31.18 |
| Energy contribution | -31.78 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1320166 111 + 20766785 CUCUAGUCCCCCCUUUC---------CACGCCUGCACCGCAAUUAUGUUUUGUGGCUUGCGGCGUGCGGACCGGAAAUUAAUUUCGUGCUUUAUUGCAACUAAUUCGAUUAUGAGUCCGC .................---------.(((((.(((((((((.......))))))..))))))))((((((((..((((((((...(((......)))...)))).)))).)).)))))) ( -34.00) >DroSec_CAF1 38700 120 + 1 CACCAGUCCCCCCAUUCCCACCAUGCCACGCCUGCACCGCAAUUAUGUUUUGUGGCUUGCGGCGUGCGGACCGGAAAUUAAUUUCGUGCUUUAUUGCAACUAAUUCGAUUAUGAGUCCGC ...........................(((((.(((((((((.......))))))..))))))))((((((((..((((((((...(((......)))...)))).)))).)).)))))) ( -34.00) >DroSim_CAF1 17783 120 + 1 CCACAGUCCCUCCAUUCCCACCAUGCCACGCCUGCACCGCAAUUAUGUUUUGUGGCUUGCGGCGUGCGGACCGGAAAUUAAUUUCGUGCUUUAUUGCAACUAAUUCGAUUAUGAGUCCGC ...........................(((((.(((((((((.......))))))..))))))))((((((((..((((((((...(((......)))...)))).)))).)).)))))) ( -34.00) >DroEre_CAF1 20168 112 + 1 ------UCCACGCCCGCC--CAAUCCCACGCCUGCACCGCAAUUAUGUUUUGUGGCUUGCGGCGUGCGCACCGGAAAUUAAUUUCGUGCUUUAUUGCAACUAAUUCGAUUAUGAGCCCGC ------.....((..((.--(((((.((((((.(((((((((.......))))))..))))))))).((((.((((.....)))))))).................)))..)).))..)) ( -29.90) >DroYak_CAF1 6193 114 + 1 ------UCCAACCGCACCUACCAUCCCCCGCCUGCACCGCAAUUAUGUUUUGUGGCUUGCGGCGUGCGGACCGGAAAUUAAUUUCGUGCUUUAUUGCAACUAAUUCGAUUAUGAGUCCGC ------......................((((.(((((((((.......))))))..))))))).((((((((..((((((((...(((......)))...)))).)))).)).)))))) ( -32.20) >consensus C___AGUCCCCCCAUUCC_ACCAUCCCACGCCUGCACCGCAAUUAUGUUUUGUGGCUUGCGGCGUGCGGACCGGAAAUUAAUUUCGUGCUUUAUUGCAACUAAUUCGAUUAUGAGUCCGC ...........................(((((.(((((((((.......))))))..))))))))((((((((..((((((((...(((......)))...)))).)))).)).)))))) (-31.18 = -31.78 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:25:20 2006