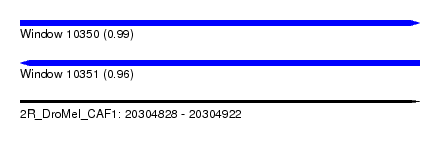
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,304,828 – 20,304,922 |
| Length | 94 |
| Max. P | 0.990914 |

| Location | 20,304,828 – 20,304,922 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 70.42 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -20.32 |
| Energy contribution | -20.85 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
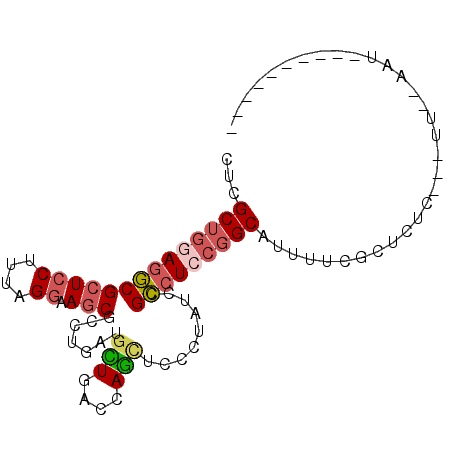
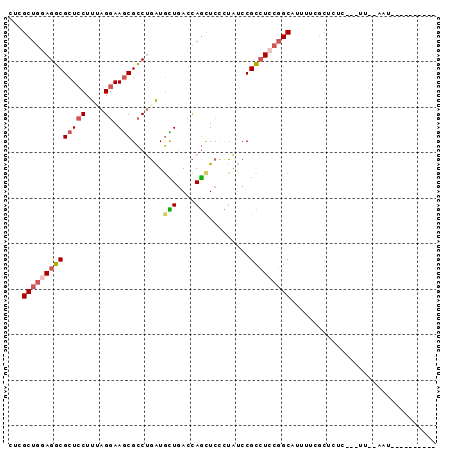
>2R_DroMel_CAF1 20304828 94 + 20766785 CUCGCUGGAGGCGCUCCUUCAGGAAGCGCCUGAUGCUGACCAGCUCCCUAUCCGCCUCCGGCAUUUUCGCUCUCGGAUCGGAACUCCUUUAUUC ...((((((((((((((....)).)))))))........)))))......((((..((((((......))...)))).))))............ ( -32.00) >DroVir_CAF1 25760 78 + 1 CGCGCUGCAGUCGCUCCUUUAGAAAACGACGCGUUUUAAUUAACUCCUGCACCGAAUCAUGCAUAAUUCAA------UUAAAAU---------- ......((.((((.((.....))...))))))(((((((((......((((........))))......))------)))))))---------- ( -11.20) >DroSec_CAF1 21721 74 + 1 CUCGCUGGAGGCGCUCCUUUAGGAAGCGCCUGAUGCUGACCAGCUCCCUAUCCGCCUCCGGCAUUUUCGCUCCU-------------------- ...((((((((((((((....)).)))....((.(((....))))).......)))))))))............-------------------- ( -28.90) >DroSim_CAF1 23417 74 + 1 CUCGCUGGAGGCGCUCCUUCAGGAAGCGCCUGAUGUUGACCAGCUCUCUAUCCGCCUCCGGCAUUUUCGCUCCU-------------------- ...((((((((((.....(((((.....))))).(((....)))........))))))))))............-------------------- ( -27.70) >DroEre_CAF1 22095 88 + 1 CUCGCUGGAGGCGCUCCUUAAGGAAGCGCCUGAUGCUGACCAGUUCCUUAUCCGCCUCCGGCAUUUGCGCUCUCGGUUUACAGUUCCU------ ...((((((((((.....(((((((....(((........))))))))))..))))))))))..........................------ ( -29.00) >DroYak_CAF1 21818 88 + 1 CCCGCUGCAGGCGCUCCUUUAGGAAGCGCCUGAUGCUGACCAGUUCCUUAUCCGCCUCCGGCAUUUGCGCUCUCAAAUUGGAAUUCCU------ ...((..((((((((((....)).))))))))..)).....((((((......(((...)))(((((......))))).))))))...------ ( -28.40) >consensus CUCGCUGGAGGCGCUCCUUUAGGAAGCGCCUGAUGCUGACCAGCUCCCUAUCCGCCUCCGGCAUUUUCGCUCUC___UU__AAU__________ ...((((((((((((((....)).))).......(((....))).........)))))))))................................ (-20.32 = -20.85 + 0.53)

| Location | 20,304,828 – 20,304,922 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 70.42 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -20.14 |
| Energy contribution | -20.62 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
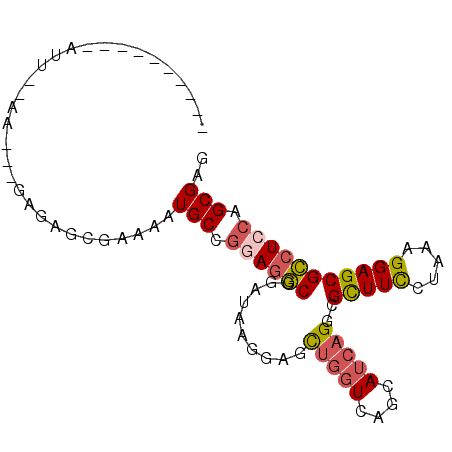
>2R_DroMel_CAF1 20304828 94 - 20766785 GAAUAAAGGAGUUCCGAUCCGAGAGCGAAAAUGCCGGAGGCGGAUAGGGAGCUGGUCAGCAUCAGGCGCUUCCUGAAGGAGCGCCUCCAGCGAG ...........((((.(((((.(.((......))).....))))).))))(((((........(((((((((.....))))))))))))))... ( -34.00) >DroVir_CAF1 25760 78 - 1 ----------AUUUUAA------UUGAAUUAUGCAUGAUUCGGUGCAGGAGUUAAUUAAAACGCGUCGUUUUCUAAAGGAGCGACUGCAGCGCG ----------.((((((------((((....(((((......)))))....)))))))))).((((((((((.....)))))))).))...... ( -22.40) >DroSec_CAF1 21721 74 - 1 --------------------AGGAGCGAAAAUGCCGGAGGCGGAUAGGGAGCUGGUCAGCAUCAGGCGCUUCCUAAAGGAGCGCCUCCAGCGAG --------------------...........(((.(((((((..((((((((..(((.......)))))))))))......))))))).))).. ( -28.70) >DroSim_CAF1 23417 74 - 1 --------------------AGGAGCGAAAAUGCCGGAGGCGGAUAGAGAGCUGGUCAACAUCAGGCGCUUCCUGAAGGAGCGCCUCCAGCGAG --------------------...........(((.((((((..........(((((....)))))..(((((.....))))))))))).))).. ( -25.70) >DroEre_CAF1 22095 88 - 1 ------AGGAACUGUAAACCGAGAGCGCAAAUGCCGGAGGCGGAUAAGGAACUGGUCAGCAUCAGGCGCUUCCUUAAGGAGCGCCUCCAGCGAG ------..................((((....)).((((((..........(((((....)))))..(((((.....))))))))))).))... ( -26.30) >DroYak_CAF1 21818 88 - 1 ------AGGAAUUCCAAUUUGAGAGCGCAAAUGCCGGAGGCGGAUAAGGAACUGGUCAGCAUCAGGCGCUUCCUAAAGGAGCGCCUGCAGCGGG ------..((.((((.(((((......))))).(((....)))....))))....)).((..((((((((((.....))))))))))..))... ( -29.10) >consensus __________AUU__AA___GAGAGCGAAAAUGCCGGAGGCGGAUAAGGAGCUGGUCAGCAUCAGGCGCUUCCUAAAGGAGCGCCUCCAGCGAG ...............................(((.((((((..........(((((....)))))..(((((.....))))))))))).))).. (-20.14 = -20.62 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:19 2006