
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,282,484 – 20,282,589 |
| Length | 105 |
| Max. P | 0.538638 |

| Location | 20,282,484 – 20,282,589 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.51 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -18.34 |
| Energy contribution | -20.40 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20282484 105 - 20766785 CAGUAGACUCCACACCAUGAAUGUGGACUUUGCGAGGGUGUGCGGGAAGCACAUAGGCGUCUACGAGGCCUACUACAAACUGGUAAGCGCACCACG--CCUCCAAGC ..(((((.((((((.......)))))).)))))((((((((((.....))))))..(((..(((.((............)).)))..)))......--))))..... ( -37.10) >DroVir_CAF1 6813 92 - 1 AAGCGGAG---GCAAUAUGAAUUUGGAUUUUGCUCGGGUGUGCGGCAAGCACAAUCUCGUCUACGAGGCCUACUAUAAACAGGUAAUG----------CGUCC--GU ..(((((.---(((.(((.......((......))((((((((.....))))...((((....))))))))...........))).))----------).)))--)) ( -27.90) >DroSec_CAF1 6180 105 - 1 CAGCGGACUCCACACCAUGAAUGUGGACUUUGCGAGGGUGUGCGGGAAGCACAUAGGCGUCUACGAGGCCUACUACAAACUGGUAAGCACACCACG--UAUCCAAGA ....(((.((((((.......))))))...((((..(((((((..........(((((.((...)).))))).(((......))).))))))).))--))))).... ( -35.50) >DroSim_CAF1 6192 105 - 1 CAGUGGACUCCACACCAUGAAUGUGGACUUUGCGAGGGUGUGCGGGAAGCACAUAGGCGUCUACGAGGCCUACUACAAACUGGUAAGCACACCACG--AAUCCAAGC ..(..((.((((((.......)))))).))..)...(((((((..........(((((.((...)).))))).(((......))).)))))))...--......... ( -34.00) >DroWil_CAF1 5908 91 - 1 ---UGA-------AAUAUGAAUUUGGAUUUUGCCAAGGUGUGCGGCAAGCACAUAGGCAUCUAUGAGGCCUACUAUAAACAGGUGAGAAU------UGAAGCAGAGG ---...-------........((..(.(((..((...((((((.....))))))((((.((...)).))))..........))..))).)------..))....... ( -22.30) >DroYak_CAF1 6221 103 - 1 CAGUGGACUCCACAUCAUGAAUGUGGACUUUGCGAGGGUGUGCGGGAAGCACAUAGGCGUCUACGAGGCCUACUACAAACUGGUAAGC--ACCACG--CCUCCAAGC ..(..((.(((((((.....))))))).))..)..((((((((.....))))))((((((....(..((.(((((.....))))).))--..))))--))))).... ( -37.20) >consensus CAGUGGACUCCACACCAUGAAUGUGGACUUUGCGAGGGUGUGCGGGAAGCACAUAGGCGUCUACGAGGCCUACUACAAACUGGUAAGCACACCACG__CAUCCAAGC ..(((((.((((((.......)))))).)))))..((((((((.....))))))((((.((...)).))))..............................)).... (-18.34 = -20.40 + 2.06)

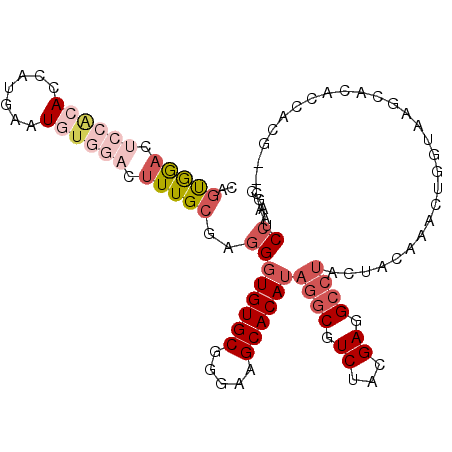
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:14 2006