| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,229,087 – 20,229,180 |
| Length | 93 |
| Max. P | 0.953313 |

| Location | 20,229,087 – 20,229,180 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.59 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
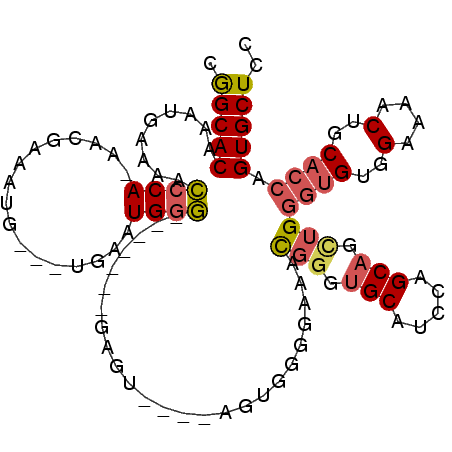
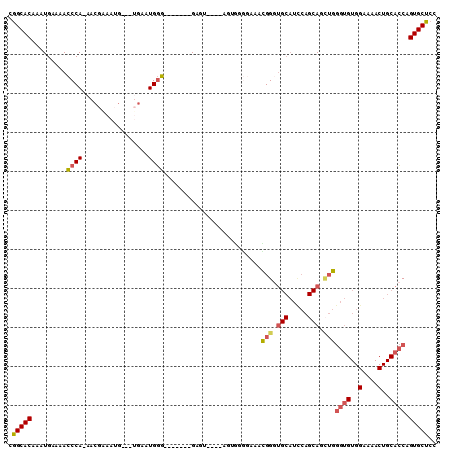
>2R_DroMel_CAF1 20229087 93 + 20766785 CGGCACAAAUGAAAACCCA-AACGGAAUGG--UGAAUGGG-------GAGC-------GGGGAAACGGAUGCAUCCAGCAGAUGGGUGUGGAAAACUGCACCAGUGCUCC .(((((.........((((-.((......)--)...))))-------..((-------(((....)....(((((((.....)))))))......))))....))))).. ( -29.20) >DroSec_CAF1 22018 100 + 1 CGGCACAAAUGAAAACCCA-AACGGAAUGC--UGAAUGGG-------GAGUGGGCAGUGGGGAAACGGGUGCAUCCAGCAGCUGGGUGUGGAAAACUGCACCAGUGCUCC .(((((.........((((-..(((....)--))..))))-------...((((((((..(....)....((((((((...)))))))).....))))).)))))))).. ( -36.90) >DroSim_CAF1 20872 100 + 1 CGGCACAAAUGAAAACCCA-AACGGAAUGG--UGAAUGGG-------GAGUGGGCAGUGGGGAAACGGGUGCAUCCAGCAGCUGGGUGUGGAAAACUGCACCAGUGCUCC .(((((.........((((-.((......)--)...))))-------...((((((((..(....)....((((((((...)))))))).....))))).)))))))).. ( -35.20) >DroEre_CAF1 23103 83 + 1 CGGCACAAAUGAA-ACCCA-AACCAAACG----GAAUGGG-------G--------------AAAUGGGUGCAUCCAGCAGCUGGGUGUGGAAAACUGCACCAGUGCUCC ..((((..((...-.((((-..((....)----)..))))-------.--------------..))..))))....((((.((((.((..(....)..)))))))))).. ( -28.90) >DroYak_CAF1 21742 92 + 1 CGGCACAAAUGAAAACCCA-AAGCAAACG----GAAUGGG-------GA------AAUGGGGAAAUGGGUGCAUUCAGCAGCUGGGUGUGGAAAACUGCACCAGUGCUCC ..((((..((.....((((-...(...(.----.....).-------).------..))))...))..))))....((((.((((.((..(....)..)))))))))).. ( -25.30) >DroAna_CAF1 20255 97 + 1 CAGCACAAAUGAAAAACCAAAACGAAAUGAAAUGAAUGCUCCGCUCCGAGU--GCAGCAUAGGAAUGG-----------AGUGGAAAGUGGAAAACUGCAGCAGUGCUCC .(((((...((......)).................((((((((((((..(--.(......).).)))-----------))))))..(..(....)..).)))))))).. ( -23.70) >consensus CGGCACAAAUGAAAACCCA_AACGAAAUG___UGAAUGGG_______GAGU____AGUGGGGAAACGGGUGCAUCCAGCAGCUGGGUGUGGAAAACUGCACCAGUGCUCC .(((((.........((((.................)))).........................(((.(((.....))).)))((((..(....)..)))).))))).. (-18.20 = -18.68 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:52 2006