
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,226,499 – 20,226,633 |
| Length | 134 |
| Max. P | 0.916817 |

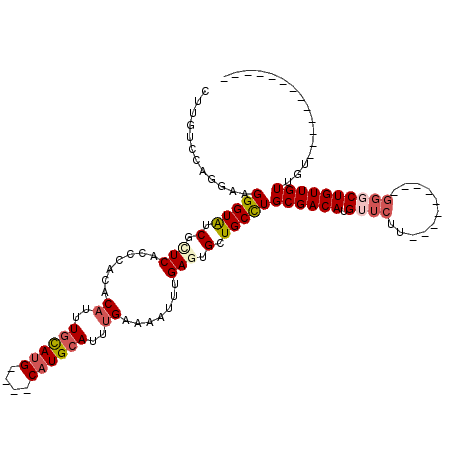
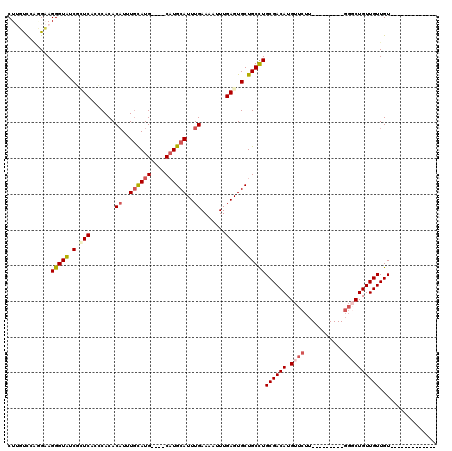
| Location | 20,226,499 – 20,226,593 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.90 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -15.70 |
| Energy contribution | -16.51 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20226499 94 - 20766785 CUUUUCCAGGAAGGGUAUCGCUCACCCACACUUUUGCAUG----CAUGCAUUUGAAAAUUUGAGUGCUGCCUGCGACAUGUUCUU---------GGGCUGUUGUUGU------------- ....((((((((((((.......))))......(((((.(----((.(((((..(....)..)))))))).)))))....)))))---------)))..........------------- ( -30.30) >DroPse_CAF1 19022 93 - 1 CU-GCUAGUGCAGGGUGUCUAUC--GCACACAUUUGUAUG----CAUGCAUUUGAAAUUUUGAGUGCUGCCUGCGACAUGUUCUU---------GGCCUGUUGUUGUUU----------- ..-((....(((((((((.....--))))(((((((((.(----((.(((((..(....)..)))))))).))))).))))....---------..)))))....))..----------- ( -27.00) >DroEre_CAF1 20554 103 - 1 CUUUUCCAGGAAGGGUAUCGCUCUGCCACACAUUUGCAUG----CAUGCAUUUGAAAAUUUGAGUGCUGCCUGCGACAUGUUCUUGCUGUUCUUGGGCUGUUGUUGU------------- ....((((((((.((((..((...))...(((((((((.(----((.(((((..(....)..)))))))).))))).))))...)))).))))))))..........------------- ( -32.00) >DroYak_CAF1 18988 94 - 1 CUUUUCCAGGAAGGGUAUCGCUCACCCACACAUUUGCAUG----CAUGCAUUUGAAAAUUUGAGUGCUGCCUGCGACAUGUUCUU---------CUGCUGUUGUUGU------------- ......(((((.((((.......))))..(((((((((.(----((.(((((..(....)..)))))))).))))).))))..))---------)))..........------------- ( -27.00) >DroAna_CAF1 17419 98 - 1 CUUGUUCCAGAAGGGUAUCCUUCACACACACAUUUAUAUGUGGGCUUGCAUUUGAAAAUUUGAGUGCUGCUUGCGACAUGUUCUU---------GGGCUGUUGUUGU------------- .........(((((....))))).....(((((....)))))(((..(((((..(....)..))))).))).((((((.(((...---------.)))))))))...------------- ( -26.80) >DroPer_CAF1 19252 104 - 1 CU-GCUAGUGCAGGGUGUCUAUC--GCACACAUUUGUAUG----CAUGCAUUUGAAAUUUUGAGUGCUGCCUGCGACAUGUUCUU---------GGCCUGUUGUUGUUUCCAUUGCCGUU ((-((....)))).((((.....--))))(((((((((.(----((.(((((..(....)..)))))))).))))).))))....---------(((.((..(....)..))..)))... ( -27.70) >consensus CUUGUCCAGGAAGGGUAUCGCUCACCCACACAUUUGCAUG____CAUGCAUUUGAAAAUUUGAGUGCUGCCUGCGACAUGUUCUU_________GGGCUGUUGUUGU_____________ ............(((((.(.(((.......((..((((((....))))))..)).......))).).)))))((((((.((((...........))))))))))................ (-15.70 = -16.51 + 0.81)

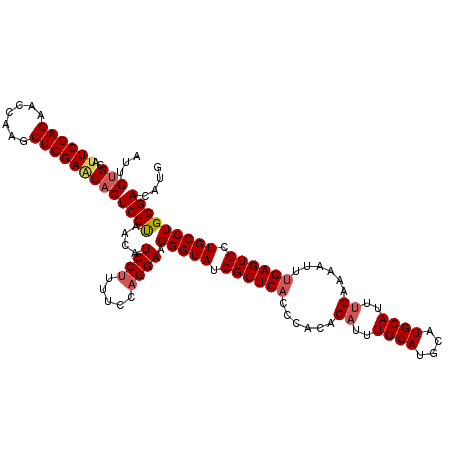
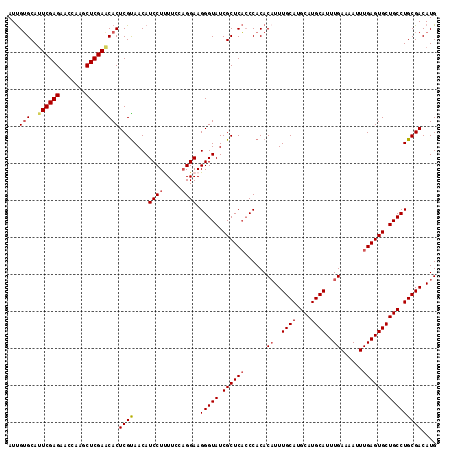
| Location | 20,226,517 – 20,226,633 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 94.05 |
| Mean single sequence MFE | -33.87 |
| Consensus MFE | -29.88 |
| Energy contribution | -30.56 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20226517 116 - 20766785 AUUGUGCAAUCGAGACUCAAGCUCGAACACUCGUAACAUCCUUUUCCAGGAAGGGUAUCGCUCACCCACACUUUUGCAUGCAUGCAUUUGAAAAUUUGAGUGCUGCCUGCGACAUG ...(((...(((((.(....)))))).)))..((....((((.....)))).((((.......))))))....(((((.(((.(((((..(....)..)))))))).))))).... ( -31.40) >DroSec_CAF1 19505 116 - 1 AUUGUGCAUUCGAGAAGCAAGCUCGAACACUCGUAACAUCCUCUUCCAGGAAGGGUAUCGCUCACCCACACAUUUGCAUGCAUGCAUUUGAAAAUUUGAGUGCUGCCUGCGACAUG ...(((..((((((.......)))))))))((((....((((.....)))).(((((.((((((......((..((((....))))..))......)))))).))))))))).... ( -33.10) >DroSim_CAF1 18367 116 - 1 AUUGUGCAUUCGAGAAGCAAGCUCGAACACUCGUAACAUCCUCUUCCUGGAAGGGUAUCGCUCACCCACACAUUUGCAUGCAUGCAUUUGAAAAUUUGAGUGCUGCCUGCGACAUG ...(((..((((((.......)))))))))((((....(((.......))).(((((.((((((......((..((((....))))..))......)))))).))))))))).... ( -32.60) >DroEre_CAF1 20581 116 - 1 AUCGAGCAUUCGAGAACCGGGCUCGAGCACUCGCAACAUCCUUUUCCAGGAAGGGUAUCGCUCUGCCACACAUUUGCAUGCAUGCAUUUGAAAAUUUGAGUGCUGCCUGCGACAUG .((((((...((.....)).))))))....((((....((((.....)))).(((((.(((((.......((..((((....))))..)).......))))).))))))))).... ( -36.64) >DroYak_CAF1 19006 116 - 1 AUUGUGCAUUCGAGAGCCAGGCUCGAGCACUCGUAACAUCCUUUUCCAGGAAGGGUAUCGCUCACCCACACAUUUGCAUGCAUGCAUUUGAAAAUUUGAGUGCUGCCUGCGACAUG ...((((..(((((.......)))))))))((((....((((.....)))).(((((.((((((......((..((((....))))..))......)))))).))))))))).... ( -35.60) >consensus AUUGUGCAUUCGAGAACCAAGCUCGAACACUCGUAACAUCCUUUUCCAGGAAGGGUAUCGCUCACCCACACAUUUGCAUGCAUGCAUUUGAAAAUUUGAGUGCUGCCUGCGACAUG ...(((..((((((.......)))))))))((((....((((.....)))).(((((.((((((......((..((((....))))..))......)))))).))))))))).... (-29.88 = -30.56 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:50 2006