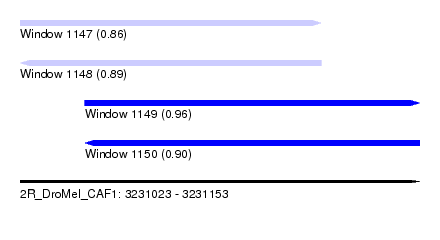
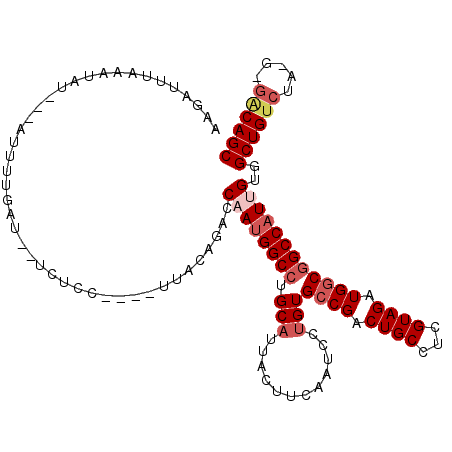
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,231,023 – 3,231,153 |
| Length | 130 |
| Max. P | 0.964289 |

| Location | 3,231,023 – 3,231,121 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.76 |
| Mean single sequence MFE | -32.54 |
| Consensus MFE | -20.31 |
| Energy contribution | -20.03 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
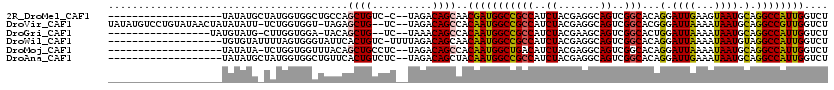
>2R_DroMel_CAF1 3231023 98 + 20766785 -------------------UAUAUGCUAUGGUGGCUGCCAGCUGUC-C--UAGACAGCAACGAUGGCCGCCAUCUACGAGGCAGUCGGCACAGGAUUGAAGUAAUGCAGGCCAUUGGUCU -------------------...((.((.((.((((((((.((((((-.--..))))))...(((((...))))).....)))))))).)).)).))...........(((((...))))) ( -35.60) >DroVir_CAF1 5985 114 + 1 UAUAUGUCCUGUAUAACUAUAUAUU-UCUGGUGGU-UAGAGCUG--UC--UAGACAGCCACAAUGGCCGCCAUCUACGAGGCAGUCGGCACGGGAUUAAAAUAAUGCAGGCCGUUGGUCU (((((.....)))))..........-..(((((((-((..((((--(.--...))))).....)))))))))......((((...((((...(.((((...)))).)..))))...)))) ( -32.30) >DroGri_CAF1 6440 97 + 1 -----------------UAUGUAUG-CUUGGUGGA-UACAGCUG--UC--UAAACAGCCACAAUGGCCGCCAUCUACGAAGCAGUCGGCACUGGAUUAAAAUAAUGCAGGCCAUUGGUCU -----------------..(((((.-(.....).)-))))((((--(.--...)))))..(((((((((((..((.......))..)))..((.((((...)))).)))))))))).... ( -31.20) >DroWil_CAF1 5190 100 + 1 -------------------UGUGUAUUUUAGUGGGUAUUCACUGUC-UUUUAGACAGCAACAAUGGCCGCCAUCUACGAGGCAGUCGGCACAGGAUUAAAAUAAUGUAGGCCAUUGGUCU -------------------((.((((((....)))))).))(((((-.....)))))...(((((((((((..((.......))..)))(((..((....))..))).)))))))).... ( -27.70) >DroMoj_CAF1 6858 98 + 1 -------------------UAUAUA-UCUGGUGGUUUACAGCUGCCUC--UAGACAGCCACAAUGGCUGACAUCUACGAGGCAGUCGGCACAGGAUUAAAAUAAUGCAGGCCAUUGGUCU -------------------......-.(..(((((((.(.((((((((--(((((((((.....)))))...)))).)))))))).)(((..............))))))))))..)... ( -37.04) >DroAna_CAF1 61 99 + 1 -------------------UAUAUGCUAUGGUGGCUGUUCACUGUCUC--UAGACAGCUACAAUGGCCGCCAUCUACGAGGCAGUCGGCACAGGAUUGAAAUAAUGCAGGCCAUUGGUCU -------------------....(((.((((((((((((..(((((..--..)))))....))))))))))))........(((((.......))))).......)))((((...)))). ( -31.40) >consensus ___________________UAUAUG_UAUGGUGGCUUACAGCUGUCUC__UAGACAGCCACAAUGGCCGCCAUCUACGAGGCAGUCGGCACAGGAUUAAAAUAAUGCAGGCCAUUGGUCU ........................................((((..........))))..(((((((((((..((.......))..)))...(.((((...)))).).)))))))).... (-20.31 = -20.03 + -0.27)
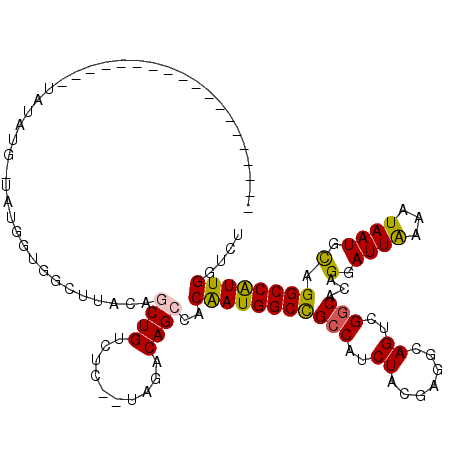
| Location | 3,231,023 – 3,231,121 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.76 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -23.32 |
| Energy contribution | -24.37 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3231023 98 - 20766785 AGACCAAUGGCCUGCAUUACUUCAAUCCUGUGCCGACUGCCUCGUAGAUGGCGGCCAUCGUUGCUGUCUA--G-GACAGCUGGCAGCCACCAUAGCAUAUA------------------- .......((((...........((....))(((((.(((..((.(((((((((((....)))))))))))--.-))))).)))))))))............------------------- ( -29.20) >DroVir_CAF1 5985 114 - 1 AGACCAACGGCCUGCAUUAUUUUAAUCCCGUGCCGACUGCCUCGUAGAUGGCGGCCAUUGUGGCUGUCUA--GA--CAGCUCUA-ACCACCAGA-AAUAUAUAGUUAUACAGGACAUAUA ...((...((((.((..............))((((.((((...)))).)))))))).(((.((((((...--.)--))))).))-)........-................))....... ( -25.24) >DroGri_CAF1 6440 97 - 1 AGACCAAUGGCCUGCAUUAUUUUAAUCCAGUGCCGACUGCUUCGUAGAUGGCGGCCAUUGUGGCUGUUUA--GA--CAGCUGUA-UCCACCAAG-CAUACAUA----------------- ....((((((((((.((((...)))).))..((((.((((...)))).)))))))))))).((((((...--.)--)))))(((-(.(.....)-.))))...----------------- ( -29.40) >DroWil_CAF1 5190 100 - 1 AGACCAAUGGCCUACAUUAUUUUAAUCCUGUGCCGACUGCCUCGUAGAUGGCGGCCAUUGUUGCUGUCUAAAA-GACAGUGAAUACCCACUAAAAUACACA------------------- ....((((((((.(((............)))((((.((((...)))).))))))))))))(..(((((.....-)))))..)...................------------------- ( -30.20) >DroMoj_CAF1 6858 98 - 1 AGACCAAUGGCCUGCAUUAUUUUAAUCCUGUGCCGACUGCCUCGUAGAUGUCAGCCAUUGUGGCUGUCUA--GAGGCAGCUGUAAACCACCAGA-UAUAUA------------------- .......(((((.(.((((...)))).).).)))).(((((((.((((...(((((.....)))))))))--)))))))(((........))).-......------------------- ( -28.50) >DroAna_CAF1 61 99 - 1 AGACCAAUGGCCUGCAUUAUUUCAAUCCUGUGCCGACUGCCUCGUAGAUGGCGGCCAUUGUAGCUGUCUA--GAGACAGUGAACAGCCACCAUAGCAUAUA------------------- ....((((((((.(((............)))((((.((((...)))).))))))))))))..((((((..--..)))))).....((.......)).....------------------- ( -29.50) >consensus AGACCAAUGGCCUGCAUUAUUUUAAUCCUGUGCCGACUGCCUCGUAGAUGGCGGCCAUUGUGGCUGUCUA__GAGACAGCUGUAAACCACCAAA_CAUAUA___________________ ....((((((((.((..............))((((.((((...)))).))))))))))))((((((((......))))))))...................................... (-23.32 = -24.37 + 1.06)

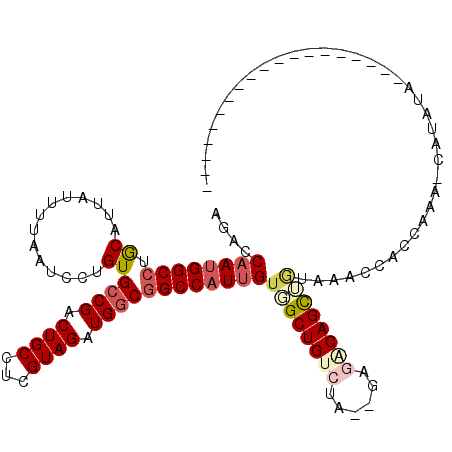
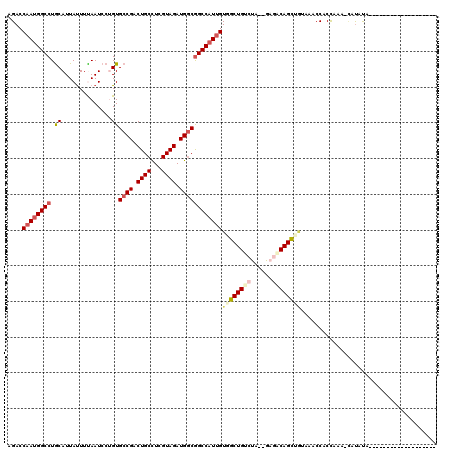
| Location | 3,231,044 – 3,231,153 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.36 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -25.17 |
| Energy contribution | -25.83 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3231044 109 + 20766785 GCUGUC-C-UAGACAGCAACGAUGGCCGCCAUCUACGAGGCAGUCGGCACAGGAUUGAAGUAAUGCAGGCCAUUGGUCUGUAA----AAAGU--UUUG-AAACAUAUGAUUAAAUCGU ((((((-.-..)))))).((((((((.(((........))).)))....(((((((.......((((((((...)))))))).----..)))--))))-..............))))) ( -32.50) >DroVir_CAF1 6023 103 + 1 GCUG--UC-UAGACAGCCACAAUGGCCGCCAUCUACGAGGCAGUCGGCACGGGAUUAAAAUAAUGCAGGCCGUUGGUCUAGAA----GGCAAAAAUGAAAAUAACAUAAG-------- (((.--((-(((((.....(((((((((((........))).....(((..(........)..))).))))))))))))))).----)))....................-------- ( -32.70) >DroPse_CAF1 17998 111 + 1 GCUGUC-CUUAGACAGCCACAAUGGCCGCCAUCUACGAGGCAGUCGGCACAGGAUUGAAGUAAUGCAGGCCAUUGGUCUGUAAUUAAAGAGA--AUCAUCAG----CAUUGGAAUCUA ((((((-....))))))..(((((((((((........)))....)))....((((....(((((((((((...))))))).)))).....)--))).....----)))))....... ( -32.40) >DroSim_CAF1 73 110 + 1 GCUGUC-C-UAGACAGCAACGAUGGCCGCCAUCUACGAGGCAGUCGGCACAGGAUUGAAGUAAUGCAGGCCAUUGGUCUGUAA----GGAGA--UUCAAAAACGCAUAUUUAAAUCGU ((((((-.-..)))))).((((((((.(((........))).))).((......(((((.(..((((((((...)))))))).----.)...--)))))....))........))))) ( -32.80) >DroMoj_CAF1 6878 113 + 1 GCUGCCUC-UAGACAGCCACAAUGGCUGACAUCUACGAGGCAGUCGGCACAGGAUUAAAAUAAUGCAGGCCAUUGGUCUGGAA----GAGACAAAUAGAGAUAGAAUAAGUAAAUCUG ((((((((-(((((((((.....)))))...)))).)))))))).(((...(.((((...)))).)..)))....((((....----.))))......((((...........)))). ( -34.20) >DroPer_CAF1 26283 111 + 1 GCUGUC-CUUAGACAGCCACAAUGGCCGCCAUCUACGAGGCAGUCGGCACAGGAUUGAAGUAAUGCAGGCCAUUGGUCUGUAAUUAAAGAGA--AUCAUCAG----CAUUGGAAUCUA ((((((-....))))))..(((((((((((........)))....)))....((((....(((((((((((...))))))).)))).....)--))).....----)))))....... ( -32.40) >consensus GCUGUC_C_UAGACAGCCACAAUGGCCGCCAUCUACGAGGCAGUCGGCACAGGAUUGAAGUAAUGCAGGCCAUUGGUCUGUAA____AGAGA__AUCAAAAA___AUAAUUAAAUCUA ((((((.....))))))..((((((((..(((.(((....(((((.......)))))..))))))..))))))))........................................... (-25.17 = -25.83 + 0.67)

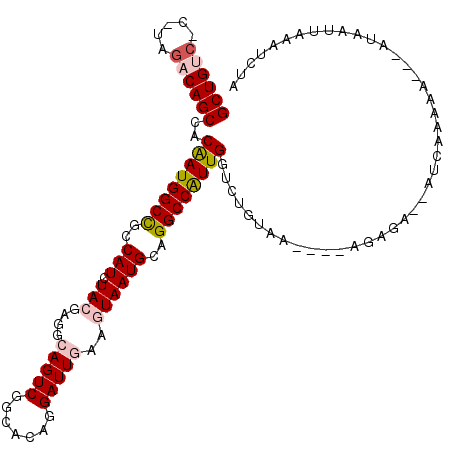
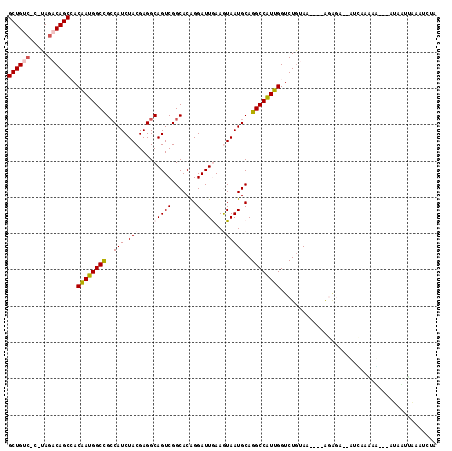
| Location | 3,231,044 – 3,231,153 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.36 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -22.66 |
| Energy contribution | -23.88 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3231044 109 - 20766785 ACGAUUUAAUCAUAUGUUU-CAAA--ACUUU----UUACAGACCAAUGGCCUGCAUUACUUCAAUCCUGUGCCGACUGCCUCGUAGAUGGCGGCCAUCGUUGCUGUCUA-G-GACAGC ...................-....--.....----.....(((..((((((.(((............)))((((.((((...)))).)))))))))).)))((((((..-.-)))))) ( -24.80) >DroVir_CAF1 6023 103 - 1 --------CUUAUGUUAUUUUCAUUUUUGCC----UUCUAGACCAACGGCCUGCAUUAUUUUAAUCCCGUGCCGACUGCCUCGUAGAUGGCGGCCAUUGUGGCUGUCUA-GA--CAGC --------....................((.----.(((((((..((((.................))))((((.((((...)))).))))((((.....)))))))))-))--..)) ( -26.23) >DroPse_CAF1 17998 111 - 1 UAGAUUCCAAUG----CUGAUGAU--UCUCUUUAAUUACAGACCAAUGGCCUGCAUUACUUCAAUCCUGUGCCGACUGCCUCGUAGAUGGCGGCCAUUGUGGCUGUCUAAG-GACAGC ............----(((.((((--(......)))))))).(((((((((.(((............)))((((.((((...)))).)))))))))))).)((((((....-)))))) ( -32.80) >DroSim_CAF1 73 110 - 1 ACGAUUUAAAUAUGCGUUUUUGAA--UCUCC----UUACAGACCAAUGGCCUGCAUUACUUCAAUCCUGUGCCGACUGCCUCGUAGAUGGCGGCCAUCGUUGCUGUCUA-G-GACAGC ..((((((((........))))))--))(((----(.((((..(.((((((.(((............)))((((.((((...)))).)))))))))).)...))))..)-)-)).... ( -32.00) >DroMoj_CAF1 6878 113 - 1 CAGAUUUACUUAUUCUAUCUCUAUUUGUCUC----UUCCAGACCAAUGGCCUGCAUUAUUUUAAUCCUGUGCCGACUGCCUCGUAGAUGUCAGCCAUUGUGGCUGUCUA-GAGGCAGC ....................(((((.((((.----....)))).))))).(.((((............)))).).(((((((.((((...(((((.....)))))))))-))))))). ( -31.50) >DroPer_CAF1 26283 111 - 1 UAGAUUCCAAUG----CUGAUGAU--UCUCUUUAAUUACAGACCAAUGGCCUGCAUUACUUCAAUCCUGUGCCGACUGCCUCGUAGAUGGCGGCCAUUGUGGCUGUCUAAG-GACAGC ............----(((.((((--(......)))))))).(((((((((.(((............)))((((.((((...)))).)))))))))))).)((((((....-)))))) ( -32.80) >consensus AAGAUUUAAAUAU___AUUUUGAU__UCUCC____UUACAGACCAAUGGCCUGCAUUACUUCAAUCCUGUGCCGACUGCCUCGUAGAUGGCGGCCAUUGUGGCUGUCUA_G_GACAGC ...........................................((((((((.(((............)))((((.((((...)))).))))))))))))..((((((.....)))))) (-22.66 = -23.88 + 1.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:37 2006