
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,215,013 – 20,215,163 |
| Length | 150 |
| Max. P | 0.882306 |

| Location | 20,215,013 – 20,215,131 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -21.62 |
| Energy contribution | -22.37 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20215013 118 + 20766785 GUUGUCAACCCGGAGUGCUCCUCCUCUUGGCCGCUUGUAAAUCGCUGGCAGACUCGGCUAAUUGGGCAACAAAAUGCUGCCCUAGUCGAAAUUUAAUUGCCUGUCAUAACUGGCCGAC ...(((.....((((.....))))....(((((.((((.....((.(((((..(((((((...(((((.(.....).)))))))))))).......))))).)).)))).)))))))) ( -36.90) >DroSec_CAF1 8141 117 + 1 GUUGUCAACCCGGAGUGCUCCUCCUCUUGGCCGCUUGUAAAUCGCUGGCAGACUCGGCUAAUUGGGCAACAAAAUGCUGCCCUAGUCGAAAUUUAAUUGCCUGUCAUAACUGG-CGAC ...(((.....((((.....)))).....(((((.........)).))).)))(((((((...(((((.(.....).)))))))))))).......(((((.((....)).))-))). ( -35.40) >DroSim_CAF1 6387 118 + 1 GUUGUCAACCCGGAGUGCUCCUCCUCUUGGCCGCUUGUAAAUCGCUGGCAGACUCGGCUAAUUGGGCAACAAAAUGCUGCCCUAGUCGAAAUUUAAUUGCCUGUCAUAACUGGCCGAC ...(((.....((((.....))))....(((((.((((.....((.(((((..(((((((...(((((.(.....).)))))))))))).......))))).)).)))).)))))))) ( -36.90) >DroEre_CAF1 9191 91 + 1 GUUGUCAACCCGGAGUGCUGCUCC---------------------------ACUCGGCUAAUUGGGCAACAAAAUGCUGCCCUAGUCGAAAUUUAAUUGCCUGUCAUAACUGGCCAAC ((((.......((((.....))))---------------------------..(((((((...(((((.(.....).)))))))))))).........(((.((....)).))))))) ( -27.50) >DroYak_CAF1 7136 91 + 1 GUUGUCAACCCGGAGUGCUCCUCC---------------------------ACUCGGCUAAUUGGGCAACAAAAUGCUGCCCUAGUCGAAAUUUAAUUGCCUGUCAUAACUGGCCAAC ((((.......((((.....))))---------------------------..(((((((...(((((.(.....).)))))))))))).........(((.((....)).))))))) ( -26.30) >DroAna_CAF1 6290 93 + 1 GUUGUCAACCCGAAGUGGCA-------------------------GGACCGACUCGGCUAAUUGGGCAACAAAAUGCUGCCCUAGUCGAAAUUUAAUUGCCUGUCAUAACUGGCCAAA .........(((..((((((-------------------------((......(((((((...(((((.(.....).))))))))))))..........))))))))...)))..... ( -27.79) >consensus GUUGUCAACCCGGAGUGCUCCUCC_____________________UGGCAGACUCGGCUAAUUGGGCAACAAAAUGCUGCCCUAGUCGAAAUUUAAUUGCCUGUCAUAACUGGCCAAC ((((.......((((.....)))).............................(((((((...(((((.(.....).)))))))))))).........(((.((....)).))))))) (-21.62 = -22.37 + 0.75)

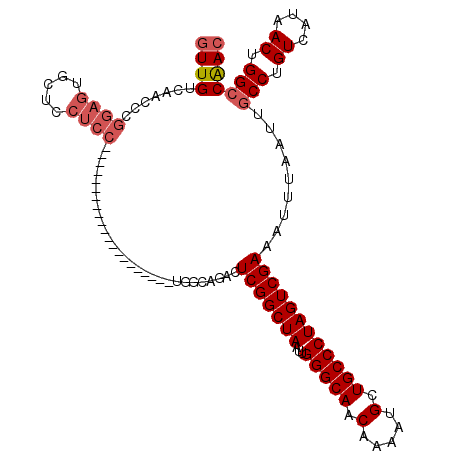
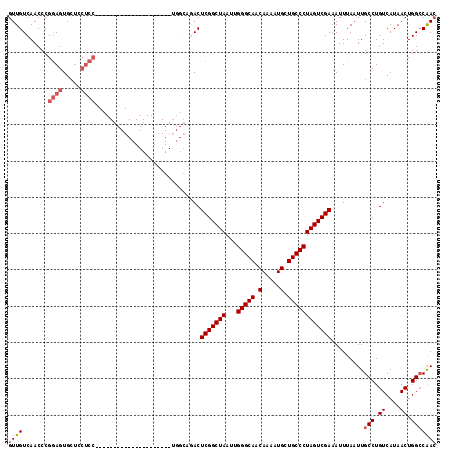
| Location | 20,215,013 – 20,215,131 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -28.82 |
| Energy contribution | -29.90 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20215013 118 - 20766785 GUCGGCCAGUUAUGACAGGCAAUUAAAUUUCGACUAGGGCAGCAUUUUGUUGCCCAAUUAGCCGAGUCUGCCAGCGAUUUACAAGCGGCCAAGAGGAGGAGCACUCCGGGUUGACAAC (((((((..........((((.......((((.((((((((((.....)))))))...))).))))..)))).((.........))))))....((((.....)))).....)))... ( -41.60) >DroSec_CAF1 8141 117 - 1 GUCG-CCAGUUAUGACAGGCAAUUAAAUUUCGACUAGGGCAGCAUUUUGUUGCCCAAUUAGCCGAGUCUGCCAGCGAUUUACAAGCGGCCAAGAGGAGGAGCACUCCGGGUUGACAAC ((((-(..((...(((.(((((......))......(((((((.....))))))).....)))..))).))..)))))......((((((....((((.....)))).))))).)... ( -38.70) >DroSim_CAF1 6387 118 - 1 GUCGGCCAGUUAUGACAGGCAAUUAAAUUUCGACUAGGGCAGCAUUUUGUUGCCCAAUUAGCCGAGUCUGCCAGCGAUUUACAAGCGGCCAAGAGGAGGAGCACUCCGGGUUGACAAC (((((((..........((((.......((((.((((((((((.....)))))))...))).))))..)))).((.........))))))....((((.....)))).....)))... ( -41.60) >DroEre_CAF1 9191 91 - 1 GUUGGCCAGUUAUGACAGGCAAUUAAAUUUCGACUAGGGCAGCAUUUUGUUGCCCAAUUAGCCGAGU---------------------------GGAGCAGCACUCCGGGUUGACAAC (((((((.((....)).))).........(((((..(((((((.....)))))))......((((((---------------------------(......)))).)))))))))))) ( -32.40) >DroYak_CAF1 7136 91 - 1 GUUGGCCAGUUAUGACAGGCAAUUAAAUUUCGACUAGGGCAGCAUUUUGUUGCCCAAUUAGCCGAGU---------------------------GGAGGAGCACUCCGGGUUGACAAC ((..(((.....................((((.((((((((((.....)))))))...))).))))(---------------------------((((.....))))))))..))... ( -32.60) >DroAna_CAF1 6290 93 - 1 UUUGGCCAGUUAUGACAGGCAAUUAAAUUUCGACUAGGGCAGCAUUUUGUUGCCCAAUUAGCCGAGUCGGUCC-------------------------UGCCACUUCGGGUUGACAAC .((((((.((....)).))).........(((((..(((((((.....)))))))......((((((.((...-------------------------..)))).)))))))))))). ( -30.50) >consensus GUCGGCCAGUUAUGACAGGCAAUUAAAUUUCGACUAGGGCAGCAUUUUGUUGCCCAAUUAGCCGAGUCUGCCA_____________________GGAGGAGCACUCCGGGUUGACAAC (((((((......(((.(((((......))......(((((((.....))))))).....)))..)))..........................((((.....)))).)))))))... (-28.82 = -29.90 + 1.08)

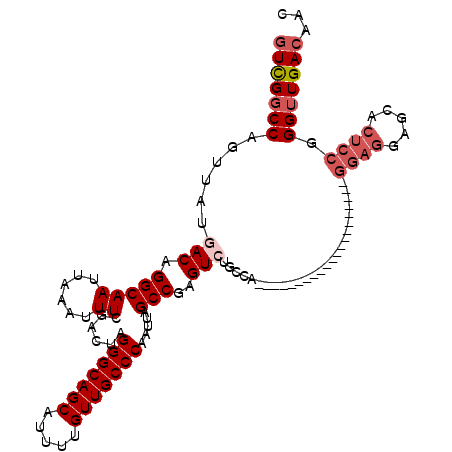
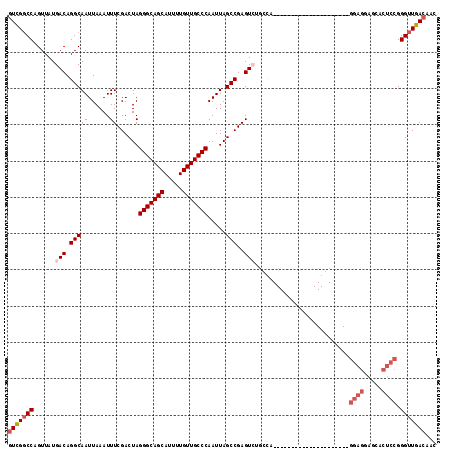
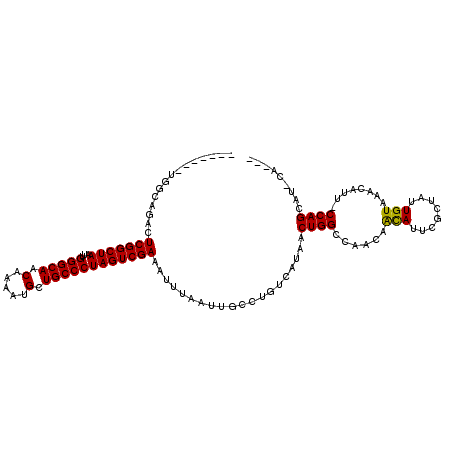
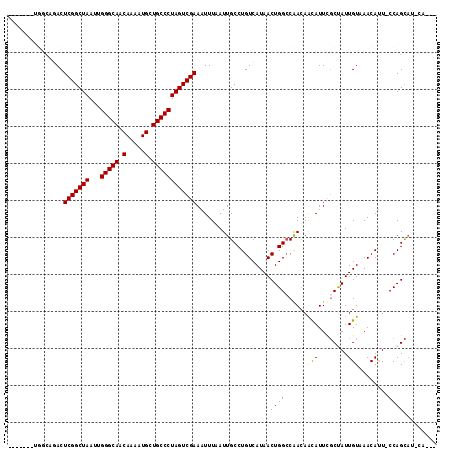
| Location | 20,215,051 – 20,215,163 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 84.02 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -19.26 |
| Energy contribution | -18.87 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20215051 112 + 20766785 AAAUCGCUGGCAGACUCGGCUAAUUGGGCAACAAAAUGCUGCCCUAGUCGAAAUUUAAUUGCCUGUCAUAACUGGCCGACAAUAUUCGCUAUUGUCAACAUU-CCAGCAUACA--- .....(((((.....(((((((...(((((.(.....).)))))))))))).........(((.((....)).))).(((((((.....)))))))......-))))).....--- ( -35.10) >DroSec_CAF1 8179 114 + 1 AAAUCGCUGGCAGACUCGGCUAAUUGGGCAACAAAAUGCUGCCCUAGUCGAAAUUUAAUUGCCUGUCAUAACUGG-CGACAAUAUUCGCUAUUGUCAACAUU-CCAGCAUACAUAC .....(((((.....(((((((...(((((.(.....).)))))))))))).........(((.((....)).))-)(((((((.....)))))))......-)))))........ ( -36.00) >DroSim_CAF1 6425 109 + 1 AAAUCGCUGGCAGACUCGGCUAAUUGGGCAACAAAAUGCUGCCCUAGUCGAAAUUUAAUUGCCUGUCAUAACUGGCCGACAAUAUUCGCUAUUGUCAACAUU-CCAGCAU------ .....(((((.....(((((((...(((((.(.....).)))))))))))).........(((.((....)).))).(((((((.....)))))))......-)))))..------ ( -35.10) >DroEre_CAF1 9215 96 + 1 -------------ACUCGGCUAAUUGGGCAACAAAAUGCUGCCCUAGUCGAAAUUUAAUUGCCUGUCAUAACUGGCCAACAACAUUCGCCAUUGUAAACAUU-CCAGCAA------ -------------..(((((((...(((((.(.....).)))))))))))).......((((.(((.((((.((((...........))))))))..)))..-...))))------ ( -21.80) >DroYak_CAF1 7160 100 + 1 -------------ACUCGGCUAAUUGGGCAACAAAAUGCUGCCCUAGUCGAAAUUUAAUUGCCUGUCAUAACUGGCCAACAACAUUCGCCAUUGUAAACAUU-CCAGCAAACU--C -------------..(((((((...(((((.(.....).)))))))))))).......((((.(((.((((.((((...........))))))))..)))..-...))))...--. ( -22.60) >DroAna_CAF1 6310 108 + 1 -------GGACCGACUCGGCUAAUUGGGCAACAAAAUGCUGCCCUAGUCGAAAUUUAAUUGCCUGUCAUAACUGGCCAAAAGCAUUUCCUAUUGUAAACACUGCCAGUUG-UAUAA -------.....((((((((((...(((((.(.....).)))))))))))).............)))(((((((((.....(((........))).......))))))))-).... ( -29.21) >consensus _______UGGCAGACUCGGCUAAUUGGGCAACAAAAUGCUGCCCUAGUCGAAAUUUAAUUGCCUGUCAUAACUGGCCAACAACAUUCGCUAUUGUAAACAUU_CCAGCAU_CA___ ...............(((((((...(((((.(.....).))))))))))))....................((((......(((........)))........))))......... (-19.26 = -18.87 + -0.39)

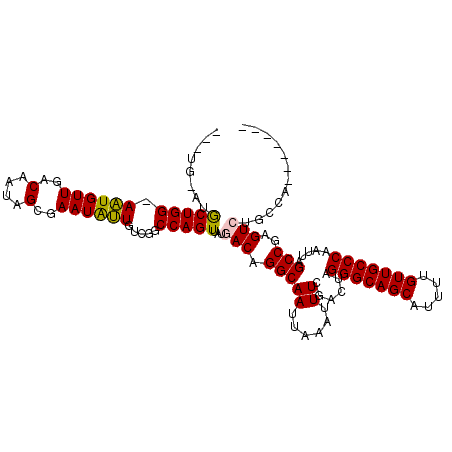
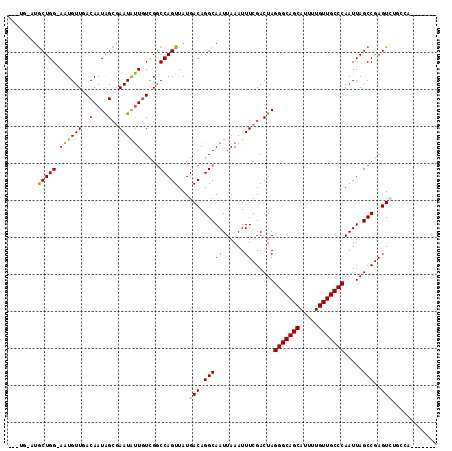
| Location | 20,215,051 – 20,215,163 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 84.02 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -23.72 |
| Energy contribution | -23.38 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20215051 112 - 20766785 ---UGUAUGCUGG-AAUGUUGACAAUAGCGAAUAUUGUCGGCCAGUUAUGACAGGCAAUUAAAUUUCGACUAGGGCAGCAUUUUGUUGCCCAAUUAGCCGAGUCUGCCAGCGAUUU ---....((((((-...((((((((((.....)))))))))).........(((((.........(((.((((((((((.....)))))))...))).)))))))))))))).... ( -39.60) >DroSec_CAF1 8179 114 - 1 GUAUGUAUGCUGG-AAUGUUGACAAUAGCGAAUAUUGUCG-CCAGUUAUGACAGGCAAUUAAAUUUCGACUAGGGCAGCAUUUUGUUGCCCAAUUAGCCGAGUCUGCCAGCGAUUU .......((((((-......(((((((.....)))))))(-((.((....)).)))........((((.((((((((((.....)))))))...))).))))....)))))).... ( -37.50) >DroSim_CAF1 6425 109 - 1 ------AUGCUGG-AAUGUUGACAAUAGCGAAUAUUGUCGGCCAGUUAUGACAGGCAAUUAAAUUUCGACUAGGGCAGCAUUUUGUUGCCCAAUUAGCCGAGUCUGCCAGCGAUUU ------.((((((-...((((((((((.....)))))))))).........(((((.........(((.((((((((((.....)))))))...))).)))))))))))))).... ( -39.60) >DroEre_CAF1 9215 96 - 1 ------UUGCUGG-AAUGUUUACAAUGGCGAAUGUUGUUGGCCAGUUAUGACAGGCAAUUAAAUUUCGACUAGGGCAGCAUUUUGUUGCCCAAUUAGCCGAGU------------- ------(((((..-..((((.....((((.(((...))).)))).....)))))))))......((((.((((((((((.....)))))))...))).)))).------------- ( -29.80) >DroYak_CAF1 7160 100 - 1 G--AGUUUGCUGG-AAUGUUUACAAUGGCGAAUGUUGUUGGCCAGUUAUGACAGGCAAUUAAAUUUCGACUAGGGCAGCAUUUUGUUGCCCAAUUAGCCGAGU------------- .--...(((((..-..((((.....((((.(((...))).)))).....)))))))))......((((.((((((((((.....)))))))...))).)))).------------- ( -30.00) >DroAna_CAF1 6310 108 - 1 UUAUA-CAACUGGCAGUGUUUACAAUAGGAAAUGCUUUUGGCCAGUUAUGACAGGCAAUUAAAUUUCGACUAGGGCAGCAUUUUGUUGCCCAAUUAGCCGAGUCGGUCC------- .....-.((((((((((((((........)))))))....))))))).((((.(((((......))......(((((((.....))))))).....)))..))))....------- ( -32.30) >consensus ___UG_AUGCUGG_AAUGUUGACAAUAGCGAAUAUUGUCGGCCAGUUAUGACAGGCAAUUAAAUUUCGACUAGGGCAGCAUUUUGUUGCCCAAUUAGCCGAGUCUGCCA_______ ........(((((.((((((..(....)..)))))).....)))))...(((.(((((......))......(((((((.....))))))).....)))..)))............ (-23.72 = -23.38 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:43 2006