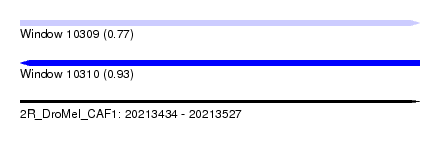
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,213,434 – 20,213,527 |
| Length | 93 |
| Max. P | 0.930370 |

| Location | 20,213,434 – 20,213,527 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 80.71 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -15.95 |
| Energy contribution | -17.31 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
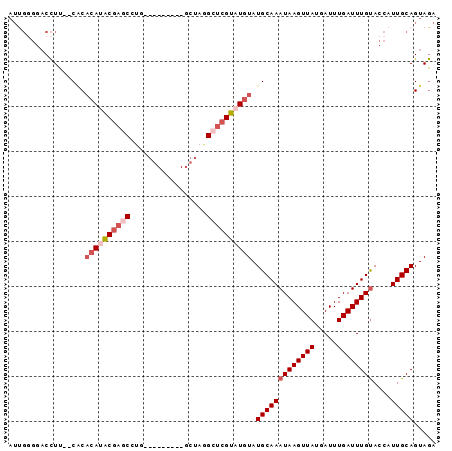
>2R_DroMel_CAF1 20213434 93 + 20766785 AUUGGGGACCUU--CACCCAUACGAGCCUGUGGCGGAGGGCUAGGCUCGUAUGUAUGCAAAUAAGUUAUGAUUUGAUUUGUACCAUUGCAGUAGA ...(((......--..)))((((((((((..(((.....)))))))))))))...(((((((((((((.....))))))))....)))))..... ( -31.20) >DroSec_CAF1 6578 93 + 1 AUUGGGGACCUU--CACACAUACGAGCCUGUGGCGCAGGGCUAGGCUCGUAUGUAUGCAAAUAAGUUAUGAUUUGAUUUGUACCAUUGCAGUAGA ...(....)...--...((((((((((((..(((.....))))))))))))))).(((((((((((((.....))))))))....)))))..... ( -31.00) >DroSim_CAF1 4771 93 + 1 AUUGGGGACCUU--CCCACAUACGAGCCUGUGGCGCAGGGCUAGGCUCGUAUGUAUGCAAAUAAGUUAUGAUUUGAUUUGUACCAUUGCAGUAGA ....(((.....--)))((((((((((((..(((.....))))))))))))))).(((((((((((((.....))))))))....)))))..... ( -35.20) >DroEre_CAF1 7618 80 + 1 AUCGAGGACCUU--CACACACACGAGCGC-------------AGGCUCGUAUGUAUGCAAAUAAGUUAUGAUUUGAUUUGCACCUUUGCAGUAGA ...((((.....--...(((.((((((..-------------..)))))).))).(((((((((((....)))).)))))))))))......... ( -21.30) >DroYak_CAF1 5486 81 + 1 AUUGGGGACCUU--CACACAUACGAACAUG------------AGGCUCGUAUGUAUGCAAAUAAGUUAUGAUUUGAUUUGUACCAUUGCAGUAGA ...(....)...--...((((((((.(...------------..).)))))))).(((((((((((((.....))))))))....)))))..... ( -17.10) >DroAna_CAF1 4581 76 + 1 -------CCUUUUUCGCAGAGGCAUAC------------GCUAGGCUCGUAUGUAUGCAAAUAAGUUAUGAUUUGAUUUGUACCAUUGCAGUAAA -------........(((...((((((------------(.......))))))).)))..((((((((.....)))))))).............. ( -15.40) >consensus AUUGGGGACCUU__CACACAUACGAGCCUG_________GCUAGGCUCGUAUGUAUGCAAAUAAGUUAUGAUUUGAUUUGUACCAUUGCAGUAGA .................((((((((((.................)))))))))).(((((((((((((.....))))))))....)))))..... (-15.95 = -17.31 + 1.36)


| Location | 20,213,434 – 20,213,527 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 80.71 |
| Mean single sequence MFE | -22.97 |
| Consensus MFE | -15.36 |
| Energy contribution | -15.72 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20213434 93 - 20766785 UCUACUGCAAUGGUACAAAUCAAAUCAUAACUUAUUUGCAUACAUACGAGCCUAGCCCUCCGCCACAGGCUCGUAUGGGUG--AAGGUCCCCAAU ..((((((((.(((...............)))...))))...(((((((((((.((.....))...)))))))))))))))--............ ( -24.46) >DroSec_CAF1 6578 93 - 1 UCUACUGCAAUGGUACAAAUCAAAUCAUAACUUAUUUGCAUACAUACGAGCCUAGCCCUGCGCCACAGGCUCGUAUGUGUG--AAGGUCCCCAAU ...........((.((....(((((........)))))(((((((((((((((.((.....))...)))))))))))))))--...)).)).... ( -27.90) >DroSim_CAF1 4771 93 - 1 UCUACUGCAAUGGUACAAAUCAAAUCAUAACUUAUUUGCAUACAUACGAGCCUAGCCCUGCGCCACAGGCUCGUAUGUGGG--AAGGUCCCCAAU .....(((((.(((...............)))...))))).((((((((((((.((.....))...))))))))))))(((--......)))... ( -28.56) >DroEre_CAF1 7618 80 - 1 UCUACUGCAAAGGUGCAAAUCAAAUCAUAACUUAUUUGCAUACAUACGAGCCU-------------GCGCUCGUGUGUGUG--AAGGUCCUCGAU .....((((....)))).....................(((((((((((((..-------------..)))))))))))))--............ ( -23.00) >DroYak_CAF1 5486 81 - 1 UCUACUGCAAUGGUACAAAUCAAAUCAUAACUUAUUUGCAUACAUACGAGCCU------------CAUGUUCGUAUGUGUG--AAGGUCCCCAAU ...........((.((....(((((........)))))(((((((((((((..------------...)))))))))))))--...)).)).... ( -20.00) >DroAna_CAF1 4581 76 - 1 UUUACUGCAAUGGUACAAAUCAAAUCAUAACUUAUUUGCAUACAUACGAGCCUAGC------------GUAUGCCUCUGCGAAAAAGG------- ..((((.....))))...............(((.((((((..((((((.......)------------)))))....)))))).))).------- ( -13.90) >consensus UCUACUGCAAUGGUACAAAUCAAAUCAUAACUUAUUUGCAUACAUACGAGCCUAGC_________CAGGCUCGUAUGUGUG__AAGGUCCCCAAU .....(((((.(((...............)))...)))))(((((((((((.................)))))))))))................ (-15.36 = -15.72 + 0.36)

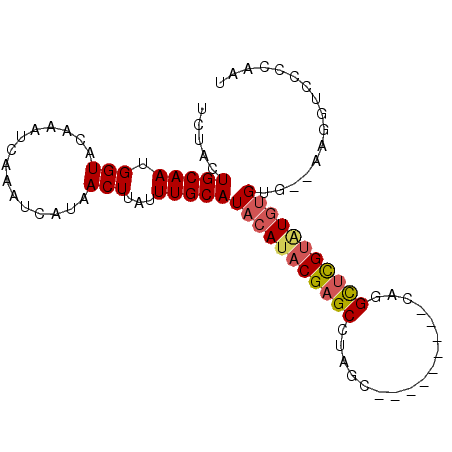

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:39 2006