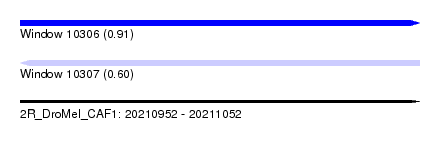
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,210,952 – 20,211,052 |
| Length | 100 |
| Max. P | 0.913562 |

| Location | 20,210,952 – 20,211,052 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.60 |
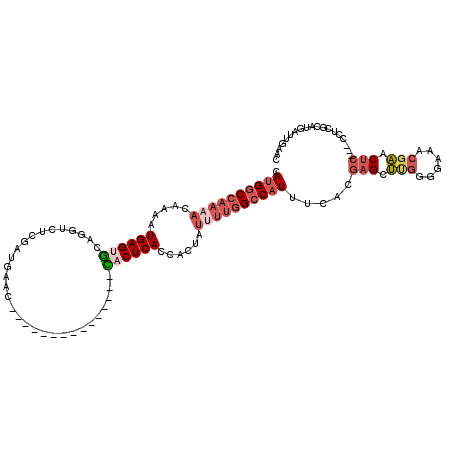
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -19.49 |
| Energy contribution | -20.96 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
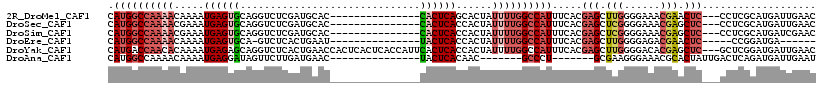
>2R_DroMel_CAF1 20210952 100 + 20766785 GUUCAAUCAUGCGAGG---GAGUUCGUUUCCCCAAGCUCGUGAAAUGGCCAAAAUAGUGCUGAGUG---------------GUGCAUCGAGACCUGCACUCAUUUUGUUUUGGCCAUG .((((.....((..((---(((.....)))))...))...))))((((((((((((.....(((((---------------(((((........))))).))))))))))))))))). ( -36.30) >DroSec_CAF1 4152 100 + 1 GUUCAAUCAUGCGAGG---GAGCUCGUUUCCCCGAGCUCGUGAAAUGGCCAAAAUAGUGGUGAGUG---------------GUGCAUCGAGACCUGCACUCAUUUCGUUUUGGCCAUG .((((.((....))..---(((((((......))))))).))))(((((((((((.(..(((((((---------------((........)))...))))))..)))))))))))). ( -37.30) >DroSim_CAF1 2367 100 + 1 GUUCGAUCAUGCGAGG---GAGCUCGUUUCCCCGAGCUCGUGAAAUGGCCAAAAUAGUGGUGAGUG---------------GUGCAUCGAGACCUGCACUCAUUUCGUUUUGGCCAUG .((((......)))).---(((((((......))))))).....(((((((((((.(..(((((((---------------((........)))...))))))..)))))))))))). ( -38.40) >DroEre_CAF1 5184 91 + 1 ------UCAUCCGG-----GAGUUCGUCUCCCCAAGCUCGUGAAAUGGCCAAAAUAGUGGUGAGUA---------------AUUCAGUGAGAC-UGCACUCAUUUUGUUUUGGCCAUG ------......((-----(((.....)))))............(((((((((((((.(((((((.---------------...(((.....)-)).)))))))))))))))))))). ( -33.50) >DroYak_CAF1 2881 115 + 1 GUUCAAUCAUCCGAGC---GAGCUCGUGUCCCCAAGCUCGUGAAAUGGCCAAAAUAGUGGUGAGUGAAUGGUGAGUGAGUGGUUCAGUGAGACCUGCUCUCAUUUUGUGUUGGUCAUG ..............((---(((((.(......).)))))))...((((((((.((((.((((((.(...(((.(.((((...)))).)...)))..).)))))))))).)))))))). ( -30.90) >DroAna_CAF1 2129 89 + 1 AUUCAAUCAUCUGAGUCAAUAGUGCGUUUCCCUUCGC-------AGGGC-------GUUGUGAGUA---------------GUUCAUCAAGAACUAUCCUCAUUUUGUUUUGGCCAUG ...........((.(((((...((((........)))-------).(((-------(..(((((((---------------((((.....))))))..)))))..))))))))))).. ( -21.90) >consensus GUUCAAUCAUCCGAGG___GAGCUCGUUUCCCCAAGCUCGUGAAAUGGCCAAAAUAGUGGUGAGUG_______________GUGCAUCGAGACCUGCACUCAUUUUGUUUUGGCCAUG ...................(((((.(......).))))).....(((((((((((...((((((((..............................))))))))..))))))))))). (-19.49 = -20.96 + 1.47)


| Location | 20,210,952 – 20,211,052 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.60 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -13.35 |
| Energy contribution | -14.88 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20210952 100 - 20766785 CAUGGCCAAAACAAAAUGAGUGCAGGUCUCGAUGCAC---------------CACUCAGCACUAUUUUGGCCAUUUCACGAGCUUGGGGAAACGAACUC---CCUCGCAUGAUUGAAC .((((((((((.....((((((...((......))..---------------))))))......))))))))))((((((.((..(((((.......))---))).)).))..)))). ( -30.00) >DroSec_CAF1 4152 100 - 1 CAUGGCCAAAACGAAAUGAGUGCAGGUCUCGAUGCAC---------------CACUCACCACUAUUUUGGCCAUUUCACGAGCUCGGGGAAACGAGCUC---CCUCGCAUGAUUGAAC .((((((((((.....((((((...((......))..---------------))))))......)))))))))).....(((((((......)))))))---................ ( -33.10) >DroSim_CAF1 2367 100 - 1 CAUGGCCAAAACGAAAUGAGUGCAGGUCUCGAUGCAC---------------CACUCACCACUAUUUUGGCCAUUUCACGAGCUCGGGGAAACGAGCUC---CCUCGCAUGAUCGAAC .((((((((((.....((((((...((......))..---------------))))))......)))))))))).....(((((((......)))))))---..(((......))).. ( -33.40) >DroEre_CAF1 5184 91 - 1 CAUGGCCAAAACAAAAUGAGUGCA-GUCUCACUGAAU---------------UACUCACCACUAUUUUGGCCAUUUCACGAGCUUGGGGAGACGAACUC-----CCGGAUGA------ .((((((((((.....((((((((-(.....)))...---------------))))))......)))))))))).........((.(((((.....)))-----)).))...------ ( -29.10) >DroYak_CAF1 2881 115 - 1 CAUGACCAACACAAAAUGAGAGCAGGUCUCACUGAACCACUCACUCACCAUUCACUCACCACUAUUUUGGCCAUUUCACGAGCUUGGGGACACGAGCUC---GCUCGGAUGAUUGAAC ...........(((..((((..(((......))).....)))).....(((((.....(((......)))........((((((((......)))))))---)...))))).)))... ( -25.00) >DroAna_CAF1 2129 89 - 1 CAUGGCCAAAACAAAAUGAGGAUAGUUCUUGAUGAAC---------------UACUCACAAC-------GCCCU-------GCGAAGGGAAACGCACUAUUGACUCAGAUGAUUGAAU ...........((...((((((((((((.....))))---------------)).)).....-------.((((-------....))))..............))))..))....... ( -17.40) >consensus CAUGGCCAAAACAAAAUGAGUGCAGGUCUCGAUGAAC_______________CACUCACCACUAUUUUGGCCAUUUCACGAGCUUGGGGAAACGAACUC___CCUCGCAUGAUUGAAC .((((((((((.....((((((..............................))))))......)))))))))).....(((.(((......))).)))................... (-13.35 = -14.88 + 1.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:36 2006