| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,210,239 – 20,210,341 |
| Length | 102 |
| Max. P | 0.540686 |

| Location | 20,210,239 – 20,210,341 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.07 |
| Mean single sequence MFE | -28.11 |
| Consensus MFE | -14.18 |
| Energy contribution | -13.84 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
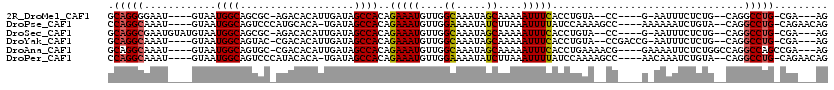
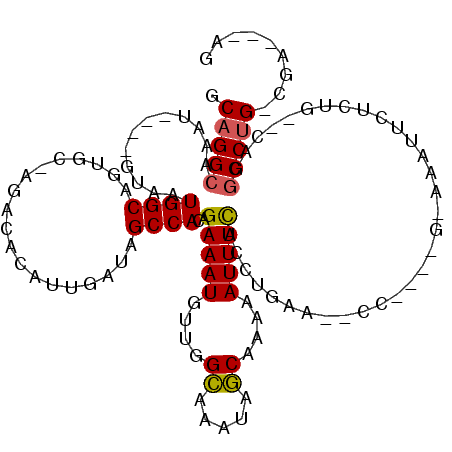
>2R_DroMel_CAF1 20210239 102 - 20766785 GCAGGGGAAU----GUAAUGGCAGCGC-AGACACAUUGAUAGCCACAGAAAUGUUGGCAAAUAGCAAAAAUUUCACCUGUA--CC----G-AAUUUCUCUG--CAGGCCUG-CGA---AG ((((((((((----.(((((.......-.....)))))...((((((....)).))))...........))))).((((((--..----(-.....)..))--))))))))-)..---.. ( -28.50) >DroPse_CAF1 1290 108 - 1 CCAGGCAAAU----GUAAUGGCAGUCCCAUGCACA-UGAUAGCCACAGAAAUGUUGGAAAAUAUCUUAAAUUUUAUCCAAAAGCC----AAAAAAUCUGUA--CAGGCCUG-CAGAACAG ...(((..((----((.((((.....))))..)))-)((((.(((((....)).)))....)))).................)))----......((((((--......))-)))).... ( -24.40) >DroSec_CAF1 3438 106 - 1 GCAGGCGAAUGUAUGUAAUGGCAGCGC-AGACACAUUGAUAGCCACAGAAAUGUUGGCAAAUAGCAAAAAUUUCACCUGUA--CC----G-AAUUUCUCUG--CAGGCCUG-CGA---AG ((((((((((((.(((....)))..))-)............((((((....)).)))).............)))..(((((--..----(-.....)..))--))))))))-)..---.. ( -29.50) >DroYak_CAF1 2106 106 - 1 GCAGGCAAAU----GUAAUGGCAGUAC-CGACACAUUGAUAGCCACAGAAAUGUUGGCAAAUAGCAAAAAUUUCACCUGUA--CCGACCG-AAUUUCUCUG--CAGGCCUG-CGA---AG ((((((.(((----((..(((.....)-))..)))))....((((((....)).))))..................(((((--..((...-....))..))--))))))))-)..---.. ( -29.80) >DroAna_CAF1 1336 108 - 1 GCAGGCAAAU----GUAAUGGCAGUGC-CGACACAUUGAUAGCCACAGAAAUGUUGGCAAAUAGCAAAAAUUUCACCUGAAAACG----GAAAAUUCUCUGGCCAGGCCAGCCGA---AG ...(((.(((----((..((((...))-))..)))))....((((((....)))((((............((((....)))).((----((......)))))))))))..)))..---.. ( -29.20) >DroPer_CAF1 1269 108 - 1 CCAGGCAAAU----GUAAUGGCAGUCCCAUACACA-UGAUAGCCACAGAAAUGUUGGAAAAUAUCUUAAAUUUUAUCCAAAAGCC----AACAAAUCUGUA--CAGGCCUG-CAGAACAG .(((((...(----(((.((((.(((.........-.))).))))((((..((((((..........................))----))))..))))))--)).)))))-........ ( -27.27) >consensus GCAGGCAAAU____GUAAUGGCAGUGC_AGACACAUUGAUAGCCACAGAAAUGUUGGCAAAUAGCAAAAAUUUCACCUGAA__CC____G_AAAUUCUCUG__CAGGCCUG_CGA___AG .(((((............((((...................))))..(((((....((.....))....)))))................................)))))......... (-14.18 = -13.84 + -0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:34 2006