| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,198,080 – 20,198,176 |
| Length | 96 |
| Max. P | 0.997323 |

| Location | 20,198,080 – 20,198,176 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.12 |
| Mean single sequence MFE | -43.40 |
| Consensus MFE | -19.06 |
| Energy contribution | -18.65 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.44 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
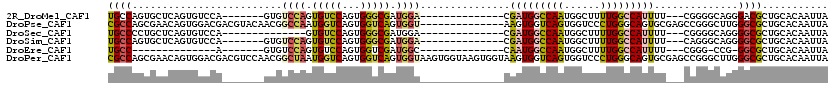
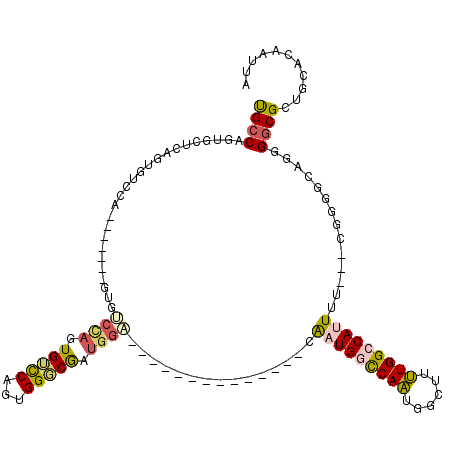
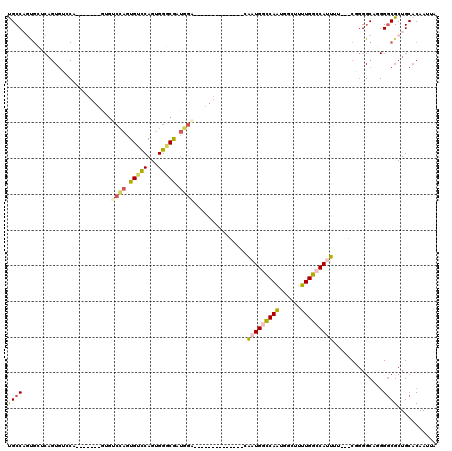
>2R_DroMel_CAF1 20198080 96 + 20766785 UGCCAGUGCUCAGUGUCCA-------GUGUCCAGUGUCCAGUGGGCGAUGGA--------------CGAUGGCCAAUGGCUUUUGGCCAUUUU---CGGGGCAGGGACGCUGCACAAUUA .....((((..(((((((.-------.(((((...(((((........))))--------------)(((((((((......)))))))))..---..))))).)))))))))))..... ( -45.40) >DroPse_CAF1 27499 106 + 1 CGCCAGCGAACAGUGGACGACGUACAACGGCCAAUGGUCAGUGGUCAGUGGU--------------AAGUGGUCAGUGGUCCCUGGGCAGUGCGAGCCGGGCUUGGGCGCUGCACAAUUA .((.((((..(((.((((.((...((...((((.((..(...)..)).))))--------------...))....)).)))))))..).((.(((((...))))).)))))))....... ( -40.30) >DroSec_CAF1 21778 89 + 1 UGCCCCUGCUCAGUGUCCA--------------GUGUCCAGUGGGCGAUGGA--------------CGAUGGCCAAUGGCUUUUGGCCAUUUU---CGGGGCAGGGGCGCUGCACAAUUA .((((((((((...(((((--------------.(((((...))))).))))--------------)(((((((((......)))))))))..---..))))))))))............ ( -49.40) >DroSim_CAF1 22106 96 + 1 UGCCAGUGCUCAGUGUCCA-------GUGUCCAGUGUCCAGUGGGCGAUGGA--------------CGAUGGCCAAUGGCUUUUGGCCAUUUU---CAGGGCAGGGGCGCUGCACAAUUA (((.(((((((..(((((.-------..(((((.(((((...))))).))))--------------)(((((((((......)))))))))..---..))))).))))))))))...... ( -48.00) >DroEre_CAF1 21755 80 + 1 UGCC--------------A-------GUGUCCAGUGUCCAGUGGUCGAUGGC--------------CAAUGGCCAAUGGCUUUUGGCCAUUUU---CGGG-CCG-GGCGCUGCACAAUUA ....--------------.-------.(((.((((((((...((((((((((--------------(((.((((...)))).)))))))))..---..))-)))-))))))).))).... ( -38.00) >DroPer_CAF1 24315 120 + 1 CGCCAGCGAACAGUGGACGACGUCCAACGGCUAAUGGUCAGUGGUCAGUGGUAAGUGGUAAGUGGUAAGUGGUCAGUGGUCCCUGGGCAGUGCGAGCCGGGCUUGGGCGCUGCACAAUUA .((.((((..((((((((...))))).(((((..((.((((.(..((.(((..(.(...........).)..))).))..).)))).)).....)))))...)))..))))))....... ( -39.30) >consensus UGCCAGUGCUCAGUGUCCA_______GUGUCCAGUGUCCAGUGGGCGAUGGA______________CAAUGGCCAAUGGCUUUUGGCCAUUUU___CGGGGCAGGGGCGCUGCACAAUUA ((((.........................((((.(((((...))))).))))...............(((((((((......)))))))))..............))))........... (-19.06 = -18.65 + -0.41)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:29 2006