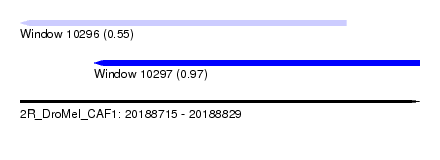
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,188,715 – 20,188,829 |
| Length | 114 |
| Max. P | 0.974296 |

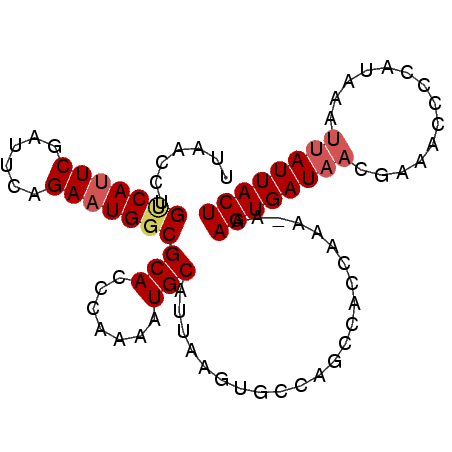
| Location | 20,188,715 – 20,188,808 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 83.12 |
| Mean single sequence MFE | -16.81 |
| Consensus MFE | -11.39 |
| Energy contribution | -12.26 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20188715 93 - 20766785 UUAACCUGUCAUUCGAUUCCGAAUGGCGCACCUGAAAUGCAUUAAGAGACAUCCACCAAAAAAAGUGAUAACGAAACCUCAUGAAUUAUUACU .......((((((((....))))))))(((.......))).......................((((((((..............)))))))) ( -15.14) >DroSim_CAF1 12820 93 - 1 UUAACCUGUCAUUCGAGUCAGAAUGGCGCACCUGAAAUGCAUUAAGGGGCACCCGCCAAAAAAAGUGAUAACGAAACCCCAUGAAUUAUUACU ....((((((((((......)))))))(((.......)))....)))(((....)))......((((((((..............)))))))) ( -21.04) >DroEre_CAF1 12540 87 - 1 UUAAGCUGCCAUUCUAUUCAGAAUGGCGCACCCAAAAUGCAUUAAGUUCCAGCCACCAAA-AAAGUGAUA-----ACGCCAUAAAUAAUUACU ....(((((((((((....))))))))(((.......)))..........))).......-..((((((.-----............)))))) ( -17.02) >DroYak_CAF1 12765 92 - 1 UUAACCUGUCAAUCGAUGCGGAAUGCCGCACCCAAAGUGCAUUAAGUUCCAGCCACCAAA-AAAGUGAUAACAAAACGCCAUAAAUUAUUACU .......(((....)))(((((((...((((.....)))).....))))).)).......-..((((((((..............)))))))) ( -14.04) >consensus UUAACCUGUCAUUCGAUUCAGAAUGGCGCACCCAAAAUGCAUUAAGUGCCAGCCACCAAA_AAAGUGAUAACGAAACCCCAUAAAUUAUUACU .......(((((((......)))))))(((.......))).......................((((((((..............)))))))) (-11.39 = -12.26 + 0.87)

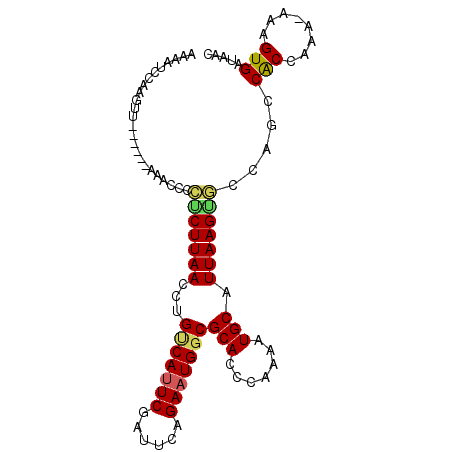
| Location | 20,188,736 – 20,188,829 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 78.96 |
| Mean single sequence MFE | -19.77 |
| Consensus MFE | -15.15 |
| Energy contribution | -15.28 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20188736 93 - 20766785 AAAUUCCAAGUU------AAAGCCCUCUUAACCUGUCAUUCGAUUCCGAAUGGCGCACCUGAAAUGCAUUAAGAGACAUCCACCAAAAAAAGUGAUAAC .........(((------(.....(((((((...((((((((....))))))))(((.......))).))))))).....(((........))).)))) ( -20.70) >DroSim_CAF1 12841 93 - 1 AAACUCCAAGUU------AAAGCCCUCUUAACCUGUCAUUCGAGUCAGAAUGGCGCACCUGAAAUGCAUUAAGGGGCACCCGCCAAAAAAAGUGAUAAC .........(((------(..((((.(((((...(((((((......)))))))(((.......))).)))))))))...(((........))).)))) ( -24.30) >DroEre_CAF1 12558 96 - 1 GAAAUUCCAGUUCUAUAAAGACCCAGCUUAAGCUGCCAUUCUAUUCAGAAUGGCGCACCCAAAAUGCAUUAAGUUCCAGCCACCAAA-AAAGUGAUA-- .........................(((..((((((((((((....))))))))(((.......)))....))))..)))(((....-...)))...-- ( -19.50) >DroYak_CAF1 12786 92 - 1 AAAACCCAAGUU------AGACACCUCUUAACCUGUCAAUCGAUGCGGAAUGCCGCACCCAAAGUGCAUUAAGUUCCAGCCACCAAA-AAAGUGAUAAC .........(((------(..(((..........(((....)))(((((((...((((.....)))).....))))).)).......-...))).)))) ( -14.60) >consensus AAAAUCCAAGUU______AAACCCCUCUUAACCUGUCAUUCGAUUCAGAAUGGCGCACCCAAAAUGCAUUAAGUGCCAGCCACCAAA_AAAGUGAUAAC ........................(((((((...(((((((......)))))))(((.......))).))))))).....(((........)))..... (-15.15 = -15.28 + 0.13)

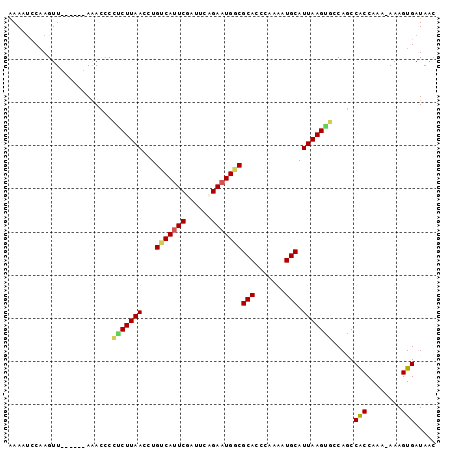
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:26 2006