| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,223,126 – 3,223,246 |
| Length | 120 |
| Max. P | 0.833530 |

| Location | 3,223,126 – 3,223,246 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.59 |
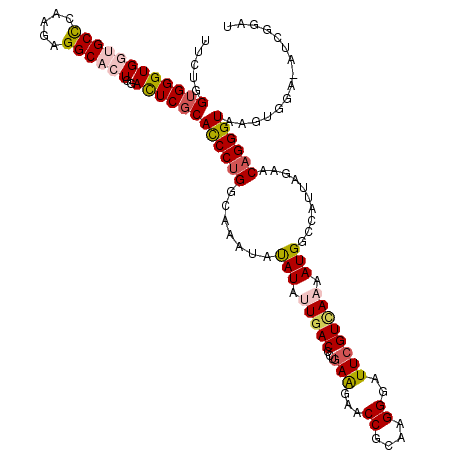
| Mean single sequence MFE | -40.52 |
| Consensus MFE | -28.15 |
| Energy contribution | -28.76 |
| Covariance contribution | 0.61 |
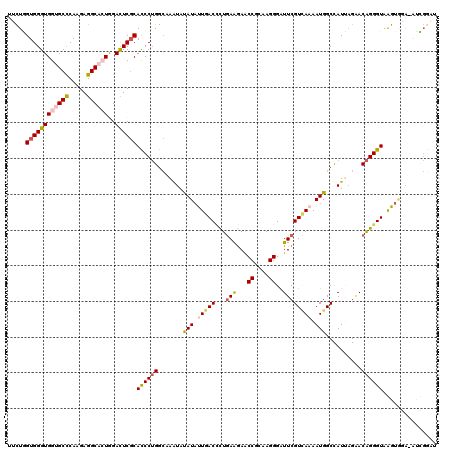
| Combinations/Pair | 1.19 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
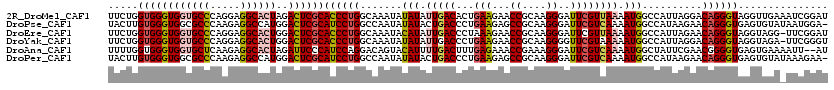
>2R_DroMel_CAF1 3223126 120 + 20766785 UUCUGGUGGGUGGUGCCCAGGAGGCACUAGACUCGCACCCUGGCAAAUAUAUAUUGACACUGAAGAACCGCAAGGGAUUCGUUAAAAUGGCCAUUAGGACAGGGUAGGUUGAAAUCGGAU .((((((((((((((((.....))))))..))))))((((((.......(((.(((((...(((...((....))..)))))))).))).((....)).))))))..........)))). ( -38.90) >DroPse_CAF1 5877 119 + 1 UACUUGUGGGUGGCGCCCAAGAGGCCAUGGACUCGCAUCCUGGCCAAUAUAUACUGACCCUGAAGAGCCGCAAGGGAUUCGUCAAAAUGGCCAUAAGAACAGGGUGAGUGUAUAAUGGA- ......((((.....))))...(((((.(((......))))))))..((((((((.((((((..(((((....))..)))(((.....)))........)))))).)))))))).....- ( -45.70) >DroEre_CAF1 5702 119 + 1 UUCUGGUGGGUGGUGCCCAGGAGGCACUGGACUCGCACCCUGGCAAAUACAUAUUGACCCUAAAGAACCGCAAGGGAUUCGUUAAAAUGGCCAUUAGAACAGGGUAGGUAGG-UUCGGAU .((((((.((.(((((((((......))))....))))))).))....((.(((..(((((...((.((....))...))(((..(((....)))..))))))))..))).)-).)))). ( -36.70) >DroYak_CAF1 5788 119 + 1 UUCUGGUGGGUGGUGCCCAGGAGGCACUGGACUCGCACCCUGGCAAAUAUAUAUUGACCCUGAAGAACCGCAAGGGGUUCGUAAAAAUGGCCAUUAGGACAGGGUAGGUAGA-UUCGGGU .((((((.((.(((((((((......))))....))))))).)).......(((..((((((.....((....))(((.(((....)))))).......))))))..)))..-..)))). ( -40.40) >DroAna_CAF1 7711 118 + 1 UUUUGGUGGGUGGUGCUCAAGAGGCACUAGAUUCCCAUCCAGGACAGUACAUUUUGACUUUGAGAAACCGAAAGGGAUUCGUCAAAAUGGCUAUUCGAACGGGGUGAGUGAAAAUU--AU (((((((((((((((((.....)))))))....)))).)))))).....(((((((((...(((...((....))..))))))))))))(((((((.....)))).))).......--.. ( -35.60) >DroPer_CAF1 5878 119 + 1 UACUUGUGGGUGGCGCCCAAGAGGCCAUGGACUCGCAUCCUGGCCAAUAUAUACUGACCCUGAAGAGCCGCAAGGGAUUCGUCAAAAUGGCCAUAAGAACAGGGUGAGUGUAUAAAGAA- ..(((.((((.....))))...(((((.(((......))))))))..((((((((.((((((..(((((....))..)))(((.....)))........)))))).)))))))))))..- ( -45.80) >consensus UUCUGGUGGGUGGUGCCCAAGAGGCACUGGACUCGCACCCUGGCAAAUAUAUAUUGACCCUGAAGAACCGCAAGGGAUUCGUCAAAAUGGCCAUUAGAACAGGGUAAGUGGA_AUCGGAU .....((((((((((((.....))))))..))))))((((((.......(((.(((((...(((...((....))..)))))))).)))..........))))))............... (-28.15 = -28.76 + 0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:32 2006