| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,124,469 – 20,124,561 |
| Length | 92 |
| Max. P | 0.663415 |

| Location | 20,124,469 – 20,124,561 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.96 |
| Mean single sequence MFE | -29.19 |
| Consensus MFE | -10.69 |
| Energy contribution | -11.70 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
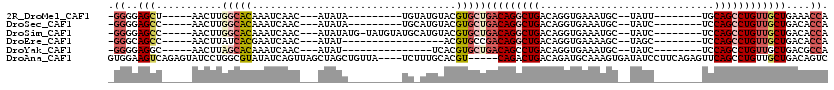
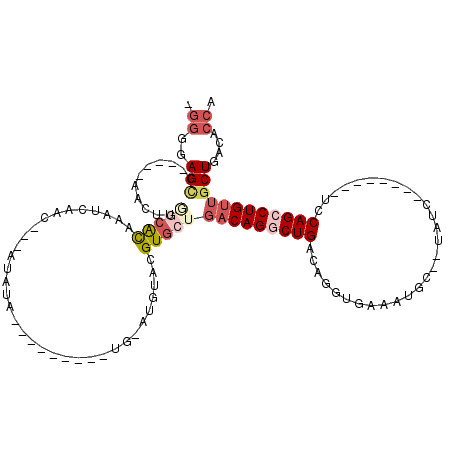
>2R_DroMel_CAF1 20124469 92 + 20766785 UGGUUUCAGCAACAGGCUGCA--------AAUA--GCAUUUCACCUGUCAGCCUGUCAGCACGUACAUACA---------UAUAU---GUUGAUUUGUGCCAAGUU-----AGCUCCCC- .((....(((.((((((((..--------.(((--(........))))))))))))..(((((.(((((..---------..)))---)).....)))))......-----.))).)).- ( -23.30) >DroSec_CAF1 9206 92 + 1 UGGUGUCAGCAACAGGCUGGA--------GAUA--GCAUUUCACCUGUCAGCCUGUCAGCACGUACAUGCA---------UAUAU---GUUGAUUUGUGCCAAGUU-----GGCUCCCC- .((.((((((.(((((((...--------((((--(........))))))))))))..(((((.(((((..---------..)))---)).....)))))...)))-----))).))..- ( -31.30) >DroSim_CAF1 9739 100 + 1 UGGUGUCAGCAACAGGCUGGA--------GAUA--GCAUUUCACCUGUCAGCCUGUCAGCACGUACAUGCAUACAUA-CAUAUAU---GUUGAUUUGUGCCAAGUU-----GGCUCCCC- .((.((((((.(((((((...--------((((--(........))))))))))))..(((((.....(((((....-....)))---)).....)))))...)))-----))).))..- ( -33.40) >DroEre_CAF1 9169 84 + 1 UGGUGUCAGCAACAGGCUGGA--------GCUA--GCUUUUCACCUGUCAGCCUGUCGGCACGU-----------------AUAU---GUUGAUUCGUGAUAAGUU-----GGCUGCCC- .(((.(((((.....)))))(--------((((--((((.((((..((((((.(((((...)).-----------------))).---))))))..)))).)))))-----))))))).- ( -32.50) >DroYak_CAF1 9146 86 + 1 UGGCGUCAGCAACAGGCUGGA--------GAUA--GCAUUUCACCUGUCAGGCUGUCAGCACGUGA---------------AUAU---GUUGAUUUGUGCUAAGUU-----GCCUCCCC- .((((.((((.(((((.((((--------(...--...))))))))))...)))).)((((((...---------------....---.......)))))).....-----))).....- ( -26.94) >DroAna_CAF1 8954 111 + 1 GACUGUCAGCAACAGGCUGAACUCUGAAGGAUAUCACUUUGCAUCUGUCAGUCUG-----ACGUGCAAAGA----UAACAGCUAGCUAACUGAUAUACGCCAGGAUACUCUGACUUCCAC ((...(((((.....)))))..)).((((.((((((((((((((..(((.....)-----)))))))))).----....((....))...))))))....((((....)))).))))... ( -27.70) >consensus UGGUGUCAGCAACAGGCUGGA________GAUA__GCAUUUCACCUGUCAGCCUGUCAGCACGUACAU_CA_________UAUAU___GUUGAUUUGUGCCAAGUU_____GGCUCCCC_ ((((((((((.(((((((((...........................)))))))))(.....).........................))))))....)))).................. (-10.69 = -11.70 + 1.00)

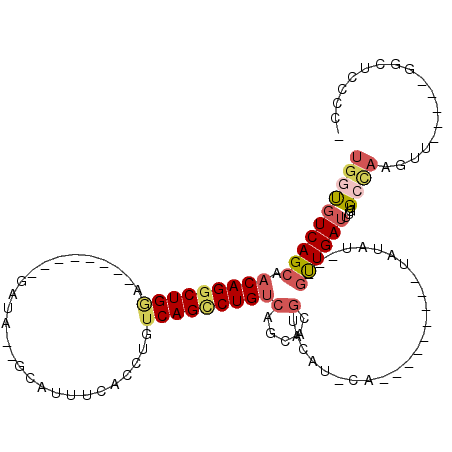
| Location | 20,124,469 – 20,124,561 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.96 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -11.13 |
| Energy contribution | -12.66 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20124469 92 - 20766785 -GGGGAGCU-----AACUUGGCACAAAUCAAC---AUAUA---------UGUAUGUACGUGCUGACAGGCUGACAGGUGAAAUGC--UAUU--------UGCAGCCUGUUGCUGAAACCA -((..(((.-----.....(((((......((---(((..---------..)))))..)))))(((((((((.((((((......--))))--------))))))))))))))....)). ( -30.70) >DroSec_CAF1 9206 92 - 1 -GGGGAGCC-----AACUUGGCACAAAUCAAC---AUAUA---------UGCAUGUACGUGCUGACAGGCUGACAGGUGAAAUGC--UAUC--------UCCAGCCUGUUGCUGACACCA -.((..(((-----.....))).....(((.(---.....---------.((((....)))).(((((((((..(((((......--))))--------).)))))))))).)))..)). ( -28.60) >DroSim_CAF1 9739 100 - 1 -GGGGAGCC-----AACUUGGCACAAAUCAAC---AUAUAUG-UAUGUAUGCAUGUACGUGCUGACAGGCUGACAGGUGAAAUGC--UAUC--------UCCAGCCUGUUGCUGACACCA -.((..(((-----.....))).....(((((---.((((((-((....)))))))).))((.(((((((((..(((((......--))))--------).))))))))))))))..)). ( -33.20) >DroEre_CAF1 9169 84 - 1 -GGGCAGCC-----AACUUAUCACGAAUCAAC---AUAU-----------------ACGUGCCGACAGGCUGACAGGUGAAAAGC--UAGC--------UCCAGCCUGUUGCUGACACCA -((.((((.-----.......((((.((....---..))-----------------.))))..(((((((((..((.((......--)).)--------).)))))))))))))...)). ( -25.40) >DroYak_CAF1 9146 86 - 1 -GGGGAGGC-----AACUUAGCACAAAUCAAC---AUAU---------------UCACGUGCUGACAGCCUGACAGGUGAAAUGC--UAUC--------UCCAGCCUGUUGCUGACGCCA -.....(((-----...(((((((........---....---------------....)))))))((((..(((((((.......--....--------....)))))))))))..))). ( -25.95) >DroAna_CAF1 8954 111 - 1 GUGGAAGUCAGAGUAUCCUGGCGUAUAUCAGUUAGCUAGCUGUUA----UCUUUGCACGU-----CAGACUGACAGAUGCAAAGUGAUAUCCUUCAGAGUUCAGCCUGUUGCUGACAGUC ......(((((......))))).....(((((.(((..((((...----.((((((((((-----(.....))).).)))))))(((......))).....))))..))))))))..... ( -30.00) >consensus _GGGGAGCC_____AACUUGGCACAAAUCAAC___AUAUA_________UG_AUGUACGUGCUGACAGGCUGACAGGUGAAAUGC__UAUC________UCCAGCCUGUUGCUGACACCA .((..(((...........(((((..................................)))))(((((((((.............................))))))))))))....)). (-11.13 = -12.66 + 1.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:15 2006