| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,122,237 – 20,122,329 |
| Length | 92 |
| Max. P | 0.937371 |

| Location | 20,122,237 – 20,122,329 |
|---|---|
| Length | 92 |
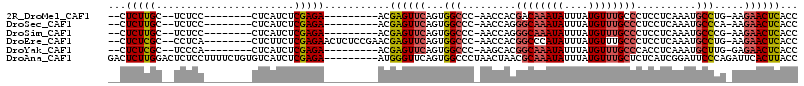
| Sequences | 6 |
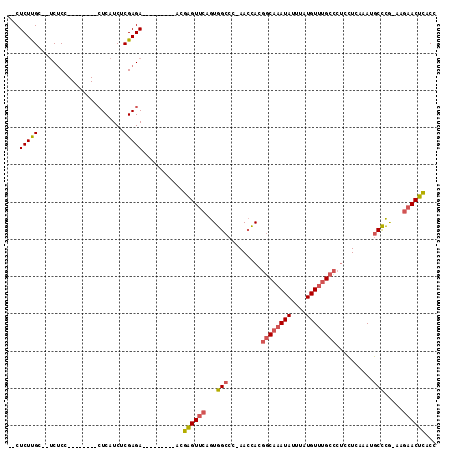
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.00 |
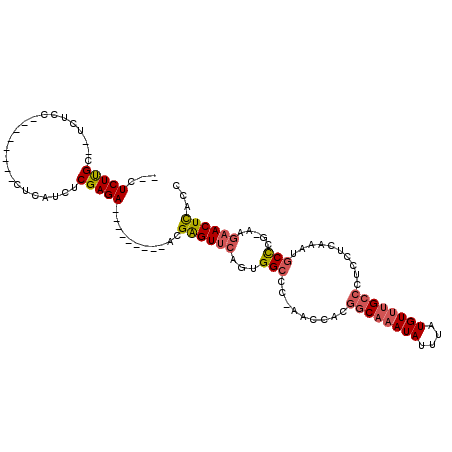
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -15.48 |
| Energy contribution | -16.01 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20122237 92 + 20766785 --CUCUUGC--UCUCC--------CUCAUCUCGAGA---------ACGAGUUCAGUGGCCC-AACCACGACAAAUAUUUAUGUUUGCCCUCCUCAAAUGCCUG-AAGAACUCACC --.....(.--((((.--------........))))---------.)((((((.((((...-..))))..((((((....)))))).................-..))))))... ( -18.20) >DroSec_CAF1 6979 92 + 1 --CUCUUGC--UCUCC--------CUCAUCUCGAGA---------ACGAGUUCAGUGGCCC-AACCAGGGCAAAUAUUUAUGUUUGCCCUCCUCAAAUGCCCA-AAGAACUCACC --.....(.--((((.--------........))))---------.)((((((..((((..-....((((((((((....))))))))))........).)))-..))))))... ( -26.74) >DroSim_CAF1 7513 92 + 1 --CUCUUGC--UCUCC--------CUCAUCUCGAGA---------ACGAGUUCAGUGGCCC-AACCAGGGCAAAUAUUUAUGUUUGCCCUCCUCAAAUGCCCG-AAGAACUCACC --.....(.--((((.--------........))))---------.)((((((.(.(((..-....((((((((((....))))))))))........)))).-..))))))... ( -27.14) >DroEre_CAF1 6980 101 + 1 --CUCUCGC--CCUCA--------CUCUUCUCGAGAACUCUCCGAACGAGUUCAGUGGCCC-AACCACGGCCCAUAUUUAUGUUUGCCCUCCUCAAAUGCCUG-AAGAACUCACC --.(((((.--.....--------.......)))))...........((((((.((((...-..))))(((.(((....)))...)))...............-..))))))... ( -20.92) >DroYak_CAF1 6948 92 + 1 --CUCUCGC--UCCCA--------CUCAUCUCGAGA---------ACGAGUUCAGUGGCCC-AAGCACGGCAAAUAUUUAUGUUUGCCCACCUCAAAUGCUUG-GAGAACUCACC --.((((..--..(((--------((...((((...---------.))))...)))))..(-(((((.((((((((....)))))))).........))))))-))))....... ( -28.90) >DroAna_CAF1 6369 106 + 1 GACUCUUGGACUCUCCUUUUCUGUGUCAUCUCGAGA---------AUGGGUUCAGUGGCCCUAACUAACGCAAAUAUUUAUGUUUGCUCUCAUCGGAUUCCCAGAUUCACUUACC ((.(((.(((...(((......(.(....).)((((---------..((((......))))........(((((((....)))))))))))...))).))).))).))....... ( -24.00) >consensus __CUCUUGC__UCUCC________CUCAUCUCGAGA_________ACGAGUUCAGUGGCCC_AACCACGGCAAAUAUUUAUGUUUGCCCUCCUCAAAUGCCCG_AAGAACUCACC ...(((((.......................)))))...........((((((...(((.........((((((((....))))))))..........))).....))))))... (-15.48 = -16.01 + 0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:14 2006