
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,117,856 – 20,117,952 |
| Length | 96 |
| Max. P | 0.603933 |

| Location | 20,117,856 – 20,117,952 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -13.13 |
| Energy contribution | -14.35 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
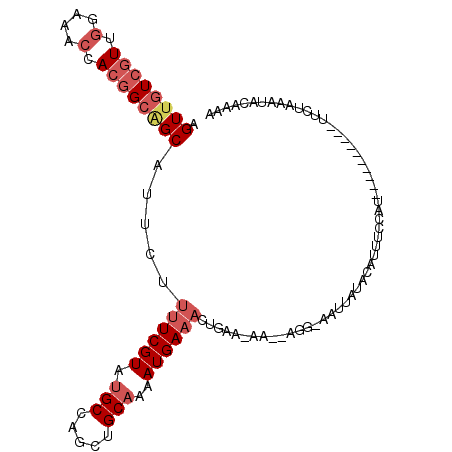
>2R_DroMel_CAF1 20117856 96 - 20766785 AGUUGUCGUUGGAAACCACGGCGGCAUUCUUUUCGUAUGCCAGCUGCAAAAUGAAACUGAAAAA--AAA-AAUUAUGCAUUUCCAU----------UUCUAAAUAGAAG- .((((((((.(....).)))))))).(((.((((((.(((.....)))..))))))..)))...--...-................----------((((....)))).- ( -20.00) >DroSec_CAF1 2599 96 - 1 AGUUGUCGUUGGAAACCACGGCAGCAUUCUUUUCGUAUGCCAGCUGCAAAAUGAAACUAAA-AA--AGG-AAUUUUACAUUUCCAU----------UUCUAGAUAGAAAA .((((((((.(....).)))))))).((((((((((.(((.....)))..))))))(((.(-((--.((-((........)))).)----------)).)))..)))).. ( -25.70) >DroSim_CAF1 3176 96 - 1 AGUUGUCGUUGGAAACCACGGCAGCAUUCUUUUCGUAUGCCAGCUGCAAAAUGAAACUAAA-AA--AGG-AAUUUUGCAUUUCCAU----------UUCUAGAUAGAAAA .((((((((.(....).)))))))).((((((((((.(((.....)))..))))))(((.(-((--.((-((........)))).)----------)).)))..)))).. ( -25.70) >DroEre_CAF1 2523 95 - 1 AGUUGUCGUUGGAAACCACGGCAGCAUUCUUUUCGUAUGCCAGCUGCAAAAUGAAACUGAA-AA--AGG-AAUUAUACAUUUUCAU-----------UCUAAAUACAAAA .((((((((.(....).)))))))).....((((((.(((.....)))..)))))).((((-((--.(.-.......).)))))).-----------............. ( -21.50) >DroYak_CAF1 2575 96 - 1 AGUUGUCGUUGGAAACCACGGCAGCAUUCUUUUCGUAUGCCAGUUGCAAAAUGAAACUGAA-AAC-AGG-AGUUAUAAAUUUUCAA-----------UCUAGAUACAAAG .((((((((.(....).)))))))).........((((..(((((.(.....).)))))..-...-.((-((........))))..-----------.....)))).... ( -21.60) >DroPer_CAF1 2692 109 - 1 AGUUAUCAUUCGACACCACAGCUGCAUUCCACUCGUACGCCAGCUGCAAUAUGAAGCUGAA-GGAUAGGAAACCAAGUAUUUUAAUGGAGAUUUAAUGGGAAAUACAAAA ...............((.((......(((((...((((..(((((.(.....).)))))..-.....(....)...)))).....)))))......)))).......... ( -16.40) >consensus AGUUGUCGUUGGAAACCACGGCAGCAUUCUUUUCGUAUGCCAGCUGCAAAAUGAAACUGAA_AA__AGG_AAUUAUACAUUUCCAU__________UUCUAAAUACAAAA .((((((((.(....).)))))))).....((((((.(((.....)))..))))))...................................................... (-13.13 = -14.35 + 1.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:12 2006