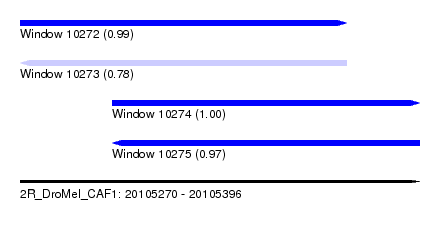
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,105,270 – 20,105,396 |
| Length | 126 |
| Max. P | 0.995544 |

| Location | 20,105,270 – 20,105,373 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.46 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -19.22 |
| Energy contribution | -19.27 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20105270 103 + 20766785 AAGUGAACAGUUCUCAA-ACUAAGCUCA-GCA-UUUGCACCGCUUCA-AUCGAGCAUUCCGACGUC---CACUGCC-AGGAUGCAACCAGAAUGCUCCUCUGGAGCCGUAC (((((...((((.....-....))))..-.))-)))..((.((((((-...((((((((.(.((((---(......-.)))))....).))))))))...)))))).)).. ( -27.40) >DroPse_CAF1 5655 96 + 1 CUUUUAACCAUUCUCAA-AGUAAGCUUA-GGACUUUGUACUGCUUCAAAUGGAGCAUUCCGAAUGC----AAUGCC-GAU--------GGAAUGCUCUUCUGGAGCCGUUC ..............(((-(((.......-..)))))).((.((((((...(((((((((((...((----...)).-..)--------))))))))))..)))))).)).. ( -28.30) >DroEre_CAF1 8088 90 + 1 -----AACAGUUCUCAA-ACUAAGCUCA-GCA-UUUGCACCGCUUCA-AUCGAGCAUUCCAACGUC---CACUGGC-GGG--------GGAAUGCUCCUCUGGAGCCGUAC -----...((((....)-))).......-((.-...))((.((((((-...(((((((((..((((---....)))-)..--------)))))))))...)))))).)).. ( -32.70) >DroYak_CAF1 8019 99 + 1 AAAUGAACAGUUCUCAA-ACUAAGCUCA-GCA-UUUGCACCGCUUCA-AUCGAGCAUUCCCACGUCAACCACUGGCAGGG--------GGAAUGCUCCUCUGGAGCCGUCC (((((...((((.....-....))))..-.))-)))..((.((((((-...((((((((((..((((.....))))...)--------)))))))))...)))))).)).. ( -32.80) >DroAna_CAF1 8567 92 + 1 AUGGCAACAGUUCUCAAAACUAAAAUUAGGCC-UUUGCACAGCUUCA-AUCGAGCAUUCUGACUCC---GA------AGU--------GGAAUGCUCCUCUGGAGCUGUCC ...((((.((..((.............))..)-)))))(((((((((-...(((((((((.(((..---..------)))--------)))))))))...))))))))).. ( -30.72) >DroPer_CAF1 5662 96 + 1 CUUUUAACCAUUCUCAA-AGUAAGCUUA-GGACUUUGUACUGCUUCAAAUGGAGCAUUCCGAAUGC----AAUGCC-GAU--------GGAAUGCUCUUCUGGAGCCGUUC ..............(((-(((.......-..)))))).((.((((((...(((((((((((...((----...)).-..)--------))))))))))..)))))).)).. ( -28.30) >consensus AUGUGAACAGUUCUCAA_ACUAAGCUCA_GCA_UUUGCACCGCUUCA_AUCGAGCAUUCCGACGUC___CACUGCC_GGG________GGAAUGCUCCUCUGGAGCCGUAC ......................................((.((((((....(((((((((............................)))))))))...)))))).)).. (-19.22 = -19.27 + 0.06)

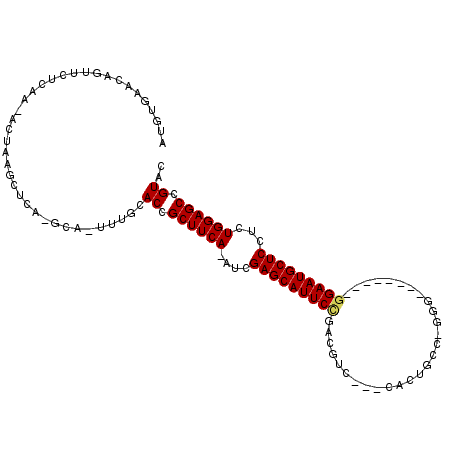
| Location | 20,105,270 – 20,105,373 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.46 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -17.05 |
| Energy contribution | -17.11 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20105270 103 - 20766785 GUACGGCUCCAGAGGAGCAUUCUGGUUGCAUCCU-GGCAGUG---GACGUCGGAAUGCUCGAU-UGAAGCGGUGCAAA-UGC-UGAGCUUAGU-UUGAGAACUGUUCACUU ((((.(((.(((..((((((((((...((.(((.-......)---)).))))))))))))..)-)).))).))))...-...-(((((..(((-(....)))))))))... ( -36.00) >DroPse_CAF1 5655 96 - 1 GAACGGCUCCAGAAGAGCAUUCC--------AUC-GGCAUU----GCAUUCGGAAUGCUCCAUUUGAAGCAGUACAAAGUCC-UAAGCUUACU-UUGAGAAUGGUUAAAAG .(((.(((.((((.(((((((((--------...-.((...----))....)))))))))..)))).)))....((((((..-.......)))-)))......)))..... ( -25.60) >DroEre_CAF1 8088 90 - 1 GUACGGCUCCAGAGGAGCAUUCC--------CCC-GCCAGUG---GACGUUGGAAUGCUCGAU-UGAAGCGGUGCAAA-UGC-UGAGCUUAGU-UUGAGAACUGUU----- ((((.(((.(((..(((((((((--------..(-(((...)---).))..)))))))))..)-)).))).))))...-...-......((((-(....)))))..----- ( -31.10) >DroYak_CAF1 8019 99 - 1 GGACGGCUCCAGAGGAGCAUUCC--------CCCUGCCAGUGGUUGACGUGGGAAUGCUCGAU-UGAAGCGGUGCAAA-UGC-UGAGCUUAGU-UUGAGAACUGUUCAUUU ((((((.((((((.(((((((((--------((..(((...)))....).))))))))))...-..((((...((...-.))-...))))..)-))).)).)))))).... ( -36.60) >DroAna_CAF1 8567 92 - 1 GGACAGCUCCAGAGGAGCAUUCC--------ACU------UC---GGAGUCAGAAUGCUCGAU-UGAAGCUGUGCAAA-GGCCUAAUUUUAGUUUUGAGAACUGUUGCCAU (.((((((.(((..((((((((.--------(((------..---..)))..))))))))..)-)).)))))).)...-(((.......((((((...))))))..))).. ( -30.00) >DroPer_CAF1 5662 96 - 1 GAACGGCUCCAGAAGAGCAUUCC--------AUC-GGCAUU----GCAUUCGGAAUGCUCCAUUUGAAGCAGUACAAAGUCC-UAAGCUUACU-UUGAGAAUGGUUAAAAG .(((.(((.((((.(((((((((--------...-.((...----))....)))))))))..)))).)))....((((((..-.......)))-)))......)))..... ( -25.60) >consensus GAACGGCUCCAGAGGAGCAUUCC________ACC_GGCAGUG___GAAGUCGGAAUGCUCGAU_UGAAGCGGUGCAAA_UGC_UAAGCUUAGU_UUGAGAACUGUUAAAAU (.((.(((.((...(((((((((............................)))))))))....)).))).)).).................................... (-17.05 = -17.11 + 0.06)

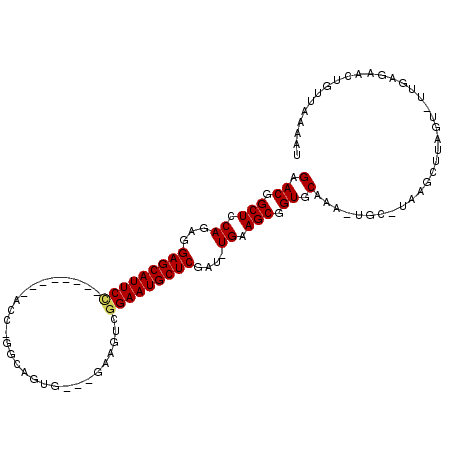
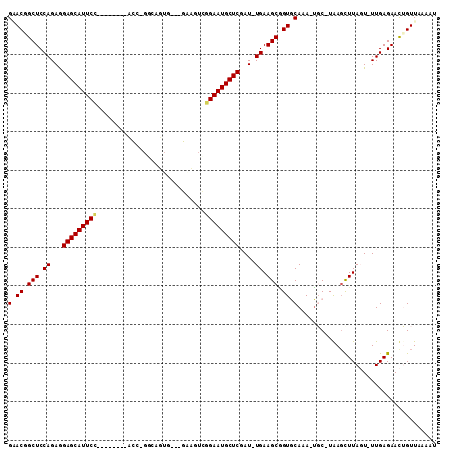
| Location | 20,105,299 – 20,105,396 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 72.91 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -20.79 |
| Energy contribution | -20.87 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20105299 97 + 20766785 A-UUUGCACCGCUUCA-AUCGAGCAUUCCGACGUCCACUGCCAGGAUGCAACCAGAAUGCUCCUCUGGAGCCGUACAAGCCACGCGAUUCAAUUCAUAC----- .-((((.((.((((((-...((((((((.(.(((((.......)))))....).))))))))...)))))).)).))))......((......))....----- ( -27.20) >DroPse_CAF1 5684 95 + 1 ACUUUGUACUGCUUCAAAUGGAGCAUUCCGAAUGC-AAUGCCGAU--------GGAAUGCUCUUCUGGAGCCGUUCAAUUUACGCGUACGCAUAGAAAUGGCAA .....((((.((((((...(((((((((((...((-...))...)--------))))))))))..))))))(((.......))).))))((.((....)))).. ( -31.10) >DroSec_CAF1 5721 97 + 1 A-UUUGCACCGCUUCA-AUCGAGCAUUCCGACGUCCACUGCCAGGAUGCUCCCAGAAUGCUCCUCUGGAGCCGUACAAGCCACGCGAUUCAAUUCAUAC----- .-((((.((.((((((-...((((((((.(((((((.......))))).))...))))))))...)))))).)).))))......((......))....----- ( -29.00) >DroEre_CAF1 8112 89 + 1 A-UUUGCACCGCUUCA-AUCGAGCAUUCCAACGUCCACUGGCGGG--------GGAAUGCUCCUCUGGAGCCGUACAAGCCACGCGAUUCAAUUCAUAC----- .-((((.((.((((((-...(((((((((..((((....))))..--------)))))))))...)))))).)).))))......((......))....----- ( -33.30) >DroAna_CAF1 8598 84 + 1 C-UUUGCACAGCUUCA-AUCGAGCAUUCUGACUCCGA-----AGU--------GGAAUGCUCCUCUGGAGCUGUCCAAUUUACGCGAAUCCAUUCAUAU----- .-.(((.(((((((((-...(((((((((.(((....-----)))--------)))))))))...))))))))).))).....................----- ( -28.90) >DroPer_CAF1 5691 95 + 1 ACUUUGUACUGCUUCAAAUGGAGCAUUCCGAAUGC-AAUGCCGAU--------GGAAUGCUCUUCUGGAGCCGUUCAAUUUACGCGUACGCAUAGAAAUGGCAA .....((((.((((((...(((((((((((...((-...))...)--------))))))))))..))))))(((.......))).))))((.((....)))).. ( -31.10) >consensus A_UUUGCACCGCUUCA_AUCGAGCAUUCCGACGUCCACUGCCAGG________GGAAUGCUCCUCUGGAGCCGUACAAGCCACGCGAAUCAAUUCAUAC_____ ...(((.((.((((((....(((((((((........((....))........)))))))))...)))))).)).))).......................... (-20.79 = -20.87 + 0.08)

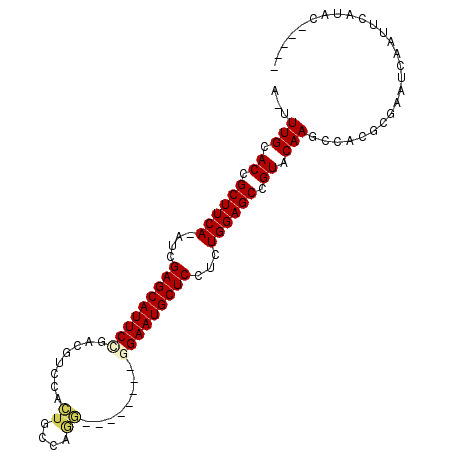
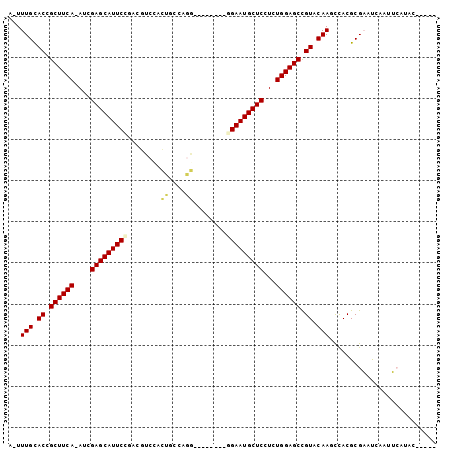
| Location | 20,105,299 – 20,105,396 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 72.91 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -18.73 |
| Energy contribution | -18.73 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20105299 97 - 20766785 -----GUAUGAAUUGAAUCGCGUGGCUUGUACGGCUCCAGAGGAGCAUUCUGGUUGCAUCCUGGCAGUGGACGUCGGAAUGCUCGAU-UGAAGCGGUGCAAA-U -----.....................((((((.(((.(((..((((((((((...((.(((.......))).))))))))))))..)-)).))).)))))).-. ( -33.20) >DroPse_CAF1 5684 95 - 1 UUGCCAUUUCUAUGCGUACGCGUAAAUUGAACGGCUCCAGAAGAGCAUUCC--------AUCGGCAUU-GCAUUCGGAAUGCUCCAUUUGAAGCAGUACAAAGU ..........(((((....)))))..(((.((.(((.((((.(((((((((--------....((...-))....)))))))))..)))).))).)).)))... ( -28.30) >DroSec_CAF1 5721 97 - 1 -----GUAUGAAUUGAAUCGCGUGGCUUGUACGGCUCCAGAGGAGCAUUCUGGGAGCAUCCUGGCAGUGGACGUCGGAAUGCUCGAU-UGAAGCGGUGCAAA-U -----.....................((((((.(((((((((.....))))))((((((.(((((.......))))).))))))...-...))).)))))).-. ( -33.30) >DroEre_CAF1 8112 89 - 1 -----GUAUGAAUUGAAUCGCGUGGCUUGUACGGCUCCAGAGGAGCAUUCC--------CCCGCCAGUGGACGUUGGAAUGCUCGAU-UGAAGCGGUGCAAA-U -----.....................((((((.(((.(((..(((((((((--------..((((...)).))..)))))))))..)-)).))).)))))).-. ( -31.40) >DroAna_CAF1 8598 84 - 1 -----AUAUGAAUGGAUUCGCGUAAAUUGGACAGCUCCAGAGGAGCAUUCC--------ACU-----UCGGAGUCAGAAUGCUCGAU-UGAAGCUGUGCAAA-G -----.....................(((.((((((.(((..((((((((.--------(((-----....)))..))))))))..)-)).)))))).))).-. ( -26.90) >DroPer_CAF1 5691 95 - 1 UUGCCAUUUCUAUGCGUACGCGUAAAUUGAACGGCUCCAGAAGAGCAUUCC--------AUCGGCAUU-GCAUUCGGAAUGCUCCAUUUGAAGCAGUACAAAGU ..........(((((....)))))..(((.((.(((.((((.(((((((((--------....((...-))....)))))))))..)))).))).)).)))... ( -28.30) >consensus _____AUAUGAAUGGAAUCGCGUAAAUUGUACGGCUCCAGAGGAGCAUUCC________ACCGGCAGUGGAAGUCGGAAUGCUCGAU_UGAAGCGGUGCAAA_U ..........................(((.((.(((.((...(((((((((........................)))))))))....)).))).)).)))... (-18.73 = -18.73 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:05 2006