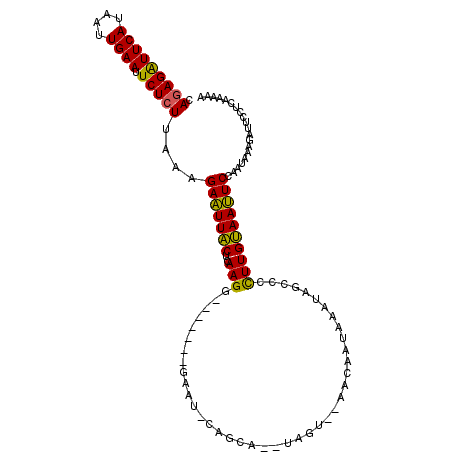
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,102,745 – 20,102,845 |
| Length | 100 |
| Max. P | 0.995064 |

| Location | 20,102,745 – 20,102,845 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.76 |
| Mean single sequence MFE | -24.14 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.09 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20102745 100 + 20766785 UUUUUGAGGAACCUUUAUUGGAAUUACAAGGGGCUAUAUAUUAUU--AAUA--UGCUAUAUUU-------CCUUGAGUAAUUCUUUAAGAGAAUUCAAUUAUGAAUCUCUG .....(((....)))....((((((((..((((..(((((.(((.--...)--)).)))))..-------))))..))))))))...(((((.((((....))))))))). ( -22.30) >DroSec_CAF1 3138 97 + 1 UUUUUGAGGAAUC--UAUUGGAAUUGCAAGGGGCUCUUUAUUGAU--ACUU--UGCUG-AUUC-------CCUUGAGCAAUUCUUAAAGAGAAUUCAAUUAUGAAUCUCUG ...(((((((.((--....((((((((((((...((......)).--.)))--))).)-))))-------)...))....)))))))(((((.((((....))))))))). ( -25.10) >DroSim_CAF1 4484 99 + 1 UUUUUGAGGAAUCUUUAUUGGAAUUACAAGGGGCUAUUUAUUAUU--ACUC--UGCUG-AUUC-------CCUUAAGUAAUUCUUUAAGAGAAUUCAAUUAUGAAUCUCUG ....((((((((.((((..(((((((..((((.............--.)))--)..))-))))-------)..))))..))))))))(((((.((((....))))))))). ( -23.44) >DroEre_CAF1 5514 105 + 1 ----UUAGGUAGCUUCAUUGGAGUUACAAAGGGCUAUU-AUGGUUUUCAGAAAGACUG-AGACAACCAAAUCUUGAGUAACUCCCUAAUAGAAUUCAAUGAUGAACCUCUG ----..((((....(((((((((((((..(((..(...-.((((((((((.....)))-)))..)))))..)))..))))))))......(....))))))...))))... ( -25.70) >consensus UUUUUGAGGAAUCUUUAUUGGAAUUACAAGGGGCUAUUUAUUAUU__AAUA__UGCUG_AUUC_______CCUUGAGUAAUUCUUUAAGAGAAUUCAAUUAUGAAUCUCUG ...................((((((((..((((.....................................))))..))))))))...(((((.((((....))))))))). (-14.90 = -14.09 + -0.81)

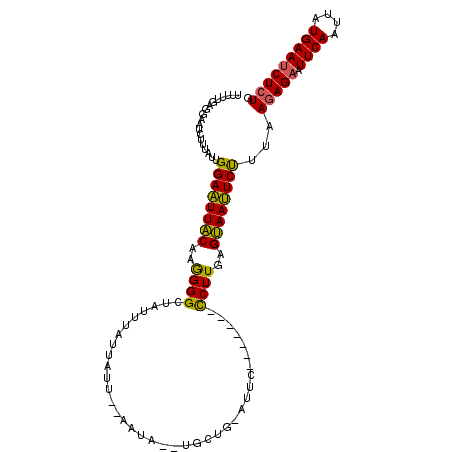
| Location | 20,102,745 – 20,102,845 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.76 |
| Mean single sequence MFE | -20.24 |
| Consensus MFE | -11.13 |
| Energy contribution | -10.25 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20102745 100 - 20766785 CAGAGAUUCAUAAUUGAAUUCUCUUAAAGAAUUACUCAAGG-------AAAUAUAGCA--UAUU--AAUAAUAUAUAGCCCCUUGUAAUUCCAAUAAAGGUUCCUCAAAAA .(((((((((....)))).)))))....(((((((....((-------..(((((..(--(...--.))..)))))..))....))))))).................... ( -17.20) >DroSec_CAF1 3138 97 - 1 CAGAGAUUCAUAAUUGAAUUCUCUUUAAGAAUUGCUCAAGG-------GAAU-CAGCA--AAGU--AUCAAUAAAGAGCCCCUUGCAAUUCCAAUA--GAUUCCUCAAAAA .(((((((((....)))).)))))....(((((((..((((-------(..(-(....--....--.........))..)))))))))))).....--............. ( -21.43) >DroSim_CAF1 4484 99 - 1 CAGAGAUUCAUAAUUGAAUUCUCUUAAAGAAUUACUUAAGG-------GAAU-CAGCA--GAGU--AAUAAUAAAUAGCCCCUUGUAAUUCCAAUAAAGAUUCCUCAAAAA .(((((((((....)))).)))))....(((((((..((((-------(...-.....--....--.............)))))))))))).................... ( -17.21) >DroEre_CAF1 5514 105 - 1 CAGAGGUUCAUCAUUGAAUUCUAUUAGGGAGUUACUCAAGAUUUGGUUGUCU-CAGUCUUUCUGAAAACCAU-AAUAGCCCUUUGUAACUCCAAUGAAGCUACCUAA---- ...((((.(.((((((....)......((((((((.....((((((((...(-(((.....)))).))))).-)))........))))))))))))).)..))))..---- ( -25.12) >consensus CAGAGAUUCAUAAUUGAAUUCUCUUAAAGAAUUACUCAAGG_______GAAU_CAGCA__UAGU__AACAAUAAAUAGCCCCUUGUAAUUCCAAUAAAGAUUCCUCAAAAA .(((((((((....)))).)))))....(((((((..(((.........................................)))))))))).................... (-11.13 = -10.25 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:00 2006