
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,097,725 – 20,097,935 |
| Length | 210 |
| Max. P | 0.993138 |

| Location | 20,097,725 – 20,097,826 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.03 |
| Mean single sequence MFE | -27.81 |
| Consensus MFE | -14.98 |
| Energy contribution | -14.74 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
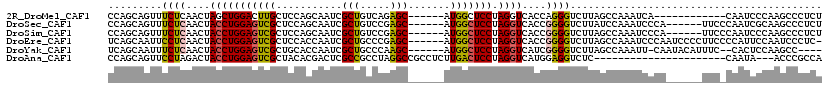
>2R_DroMel_CAF1 20097725 101 - 20766785 CCAGCAGUUUCUCAACUAGCUGGACUUGCUCCAGCAAUCGCUGUCAGAGC------AUGGCUCCUAGGUCACCAGGGUCUUAGCCAAAUCA------------CAAUCCCAAGCCCUCU (((((((((....)))).)))))...(((((((((....))))...))))------)(((((...((..(.....)..)).))))).....------------................ ( -30.70) >DroSec_CAF1 11985 107 - 1 CCAGCAGUUUCUCAACUACCUGGAGUCGCUCCAGCAAUCGCUGUCCGAGC------AUGGCUCCUAGGUCACCGGGGUCUUAUCCAAAUCCCA------UUCCCAAUCGCAAGCCCUCU ...((............((((((((((((((((((....))))...))))------..)))))).))))....(((((.........))))).------.........))......... ( -30.90) >DroSim_CAF1 12543 107 - 1 CCAGCAGUUUCUCAACUACCUGGAGUCGCUCCAGCAAUCGCUGUCCGAGC------AUGGCUCCUAGGUCACCGGGGUCUUAGCCAAAUCCCA------UUCCCAAUCCCAAGCCCUCU ...((............((((((((((((((((((....))))...))))------..)))))).))))....(((((.........))))).------.............))..... ( -29.90) >DroEre_CAF1 12172 112 - 1 UCAGCAAUUCCUCAACUACCUGGAGUCGCUCCACCAAUCGCUGCCCGAGC------AUGGCUCCUAGGUCACCGGGGUCUUAGCCAAAUCCCAAUCCCCUUCCCCAUUCCAAUCCCUC- ...((((..((((....((((((((((((((...((.....))...))))------..)))))).))))....))))..)).))..................................- ( -24.50) >DroYak_CAF1 12987 106 - 1 UCAGCAAUUUCUCAACUACCUGGAGUCGCUGCACCAAUCGCUGCCCAAGC------AUGGCUCCUAGGUCAUCGGGGUCUUAGCCAAAUU-CAAUACAUUUC--CACUCCAAGCC---- ...((............((((((((((((((((.(....).)))...)))------..)))))).))))....(((((............-...........--.)))))..)).---- ( -22.26) >DroAna_CAF1 12900 94 - 1 CCAGCAGUUCCUAGACUACCUGGAGUCGCUACACGACUCGCCGCCUAGGCCGCCUCUUGACUCCUAGGUCAUGGAGGUCUC----------------------CAAUA---ACCCGCCA .(((.((((....))))..)))((((((.....))))))(((.....))).(((((((((((....))))).))))))...----------------------.....---........ ( -28.60) >consensus CCAGCAGUUUCUCAACUACCUGGAGUCGCUCCACCAAUCGCUGCCCGAGC______AUGGCUCCUAGGUCACCGGGGUCUUAGCCAAAUCCCA______UUC_CAAUCCCAAGCCCUCU .........((((....(((((((((((...........(((.....))).......))))))).))))....)))).......................................... (-14.98 = -14.74 + -0.25)
| Location | 20,097,826 – 20,097,935 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 73.78 |
| Mean single sequence MFE | -39.47 |
| Consensus MFE | -22.50 |
| Energy contribution | -24.78 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20097826 109 - 20766785 CCUAA----AGAAAUCACAAGGAUUUCA---ACCCGGUGCAGCGGAAGUGGAUAUUCGACAGUCGCUGCUCCUGCCACCGAAAACCAGGUGGCAACCAGGGUCAGUUCUUCACCUA .....----.((((((.....)))))).---((((((.(((((((..((.(.....).))..))))))).))(((((((........)))))))....)))).............. ( -43.70) >DroVir_CAF1 22284 113 - 1 UUACCCUUCCAGCUGCCCGAGUACGUGG---UGCCGGAGCAAAGGCGCUGGCUCUAUGACGAUAAGUGUCCCUAUCAUCAUGAUCUCAAGUGCAGCCAGCGUCAGUUCUUUACCUA ..........(((((((((.((((...)---)))))).))...((((((((((((((((.((((........)))).))))).........).))))))))))))))......... ( -34.00) >DroSec_CAF1 12092 109 - 1 UCUUU----AGCCUGGUCAAGGAUAUCA---ACCCAAUCCAGCGGAAGUGGAUAUUCGACAGUCGCUGCUCCUACCACCGAAAACCAGGUGGCAACCAGGGUCAGUUCUUCACCUA .....----.....(((.((((((....---((((....((((((..((.(.....).))..))))))......(((((........)))))......))))..)))))).))).. ( -33.40) >DroSim_CAF1 12650 109 - 1 UCUUC----AGCCUGGUCAAGGAUAUCA---ACCCGGUGCAGCGGAAGUGGAUAUUCGACAGUCGCUGCUCCUACCACCGAAAACCAGGUGGCAACCAGGGUCAGUUCUUCACCUA .....----.....(((.((((((....---((((((.(((((((..((.(.....).))..))))))).))..(((((........)))))......))))..)))))).))).. ( -41.20) >DroEre_CAF1 12284 112 - 1 UCAUC----AGCCUGGUCAAGGAAUUAAGCGCCCCGGUGCAGCGGAAGUGGAUAUUCGACAGUCGCUGCUCCUACCACCGAAACCCAGGUGGAAACCAGGGUCAGUUCUUCACCUA .....----.....(((..(((((((....((((.((((((((((..((.(.....).))..))))))).....(((((........)))))..))).)))).))))))).))).. ( -40.50) >DroYak_CAF1 13093 106 - 1 UCGUC----GGCCUGG------AUCGCAGGCCCCCUGUGCAGCGGAAGUGGAUAUUCGACAGUCGCUGCUCCUACCACCGCCAACCAGGCGGAAACCAGGGUCAGUUCUUCACCUA .....----((((((.------....))))))(((((.(((((((..((.(.....).))..)))))))........(((((.....)))))....)))))............... ( -44.00) >consensus UCUUC____AGCCUGGUCAAGGAUAUCA___ACCCGGUGCAGCGGAAGUGGAUAUUCGACAGUCGCUGCUCCUACCACCGAAAACCAGGUGGCAACCAGGGUCAGUUCUUCACCUA ...........((((((..................((.(((((((..((.(.....).))..))))))).))..(((((........)))))..))))))................ (-22.50 = -24.78 + 2.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:57 2006