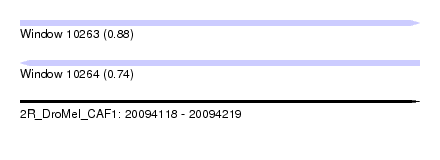
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,094,118 – 20,094,219 |
| Length | 101 |
| Max. P | 0.880742 |

| Location | 20,094,118 – 20,094,219 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.13 |
| Mean single sequence MFE | -13.26 |
| Consensus MFE | -10.88 |
| Energy contribution | -10.35 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
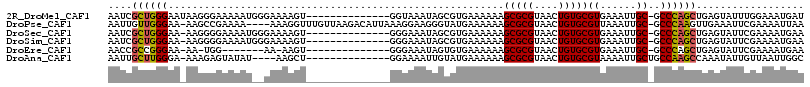
>2R_DroMel_CAF1 20094118 101 + 20766785 AUCAUUUCCAAAUACUCAGCUGGGC-GCAAUUUCACGCACAGUUACGCGCUUUUUUCACGCUAUUUACC--------------ACUUUUCCCAUUUUUCCCUUAUUCCCAGCGAUU ..................((((((.-((........))........(((.........)))........--------------.......................)))))).... ( -15.30) >DroPse_CAF1 9124 110 + 1 UUAAUUUUCGAAUUUCAACUUGGGC-GCAAUUUAACGCACAGUUACGCGCUUUUUUCAUACCCUUCCUUUAAUGUCUUAACAAACCUUU----UUUUCGGCUU-UUCCCAACAAUU .......(((((.........((((-((....((((.....)))).))))))....................(((....))).......----..)))))...-............ ( -11.20) >DroSec_CAF1 8382 100 + 1 UUCAUUUUCGAAUACUCAGCUGGGC-GCAAUUUCACGCACAGUUACGCGCUUUUUUCACGCUAUUUCCC--------------ACUUUUCCCAUUUUCCCCUU-UUCCCAGCGAUU ..................((((((.-((........))........(((.........)))........--------------....................-..)))))).... ( -15.30) >DroSim_CAF1 8362 100 + 1 UUCAUUUUCGAAUACUCAGCUGGGC-GCAAUUUCACGCACAGUUACGCGCUUUUUUCACGCUAUUUCCC--------------ACUUUUCCCAUUUUCCCCUU-UUCCCAGCGAUU ..................((((((.-((........))........(((.........)))........--------------....................-..)))))).... ( -15.30) >DroEre_CAF1 8396 91 + 1 UUCAUUUUCGAAUACUCAGCUGGGC-GCAAUUUCACGCACAGUUACGCGCUUUUUUCACACUAUUUCCC--------------ACUU-UU-------CCA-UU-UUCCCGGCGGUU ..................((((((.-((........))..(((.....)))..................--------------....-..-------...-..-..)))))).... ( -11.60) >DroAna_CAF1 9017 97 + 1 GCCAAUUAACAAUAUUUGGCUUGGCAGCAAUUUUACGCACAGUUACGCGCUUUUUUCAUACAAUUUUCC--------------AGCUU----AUAUACUCUUU-UCCCAAGCAAUU ..................((((((............((........))(((..................--------------)))..----...........-..)))))).... ( -10.87) >consensus UUCAUUUUCGAAUACUCAGCUGGGC_GCAAUUUCACGCACAGUUACGCGCUUUUUUCACACUAUUUCCC______________ACUUUUC__AUUUUCCCCUU_UUCCCAGCGAUU ..................((((((..((........))..(((.....))).......................................................)))))).... (-10.88 = -10.35 + -0.53)

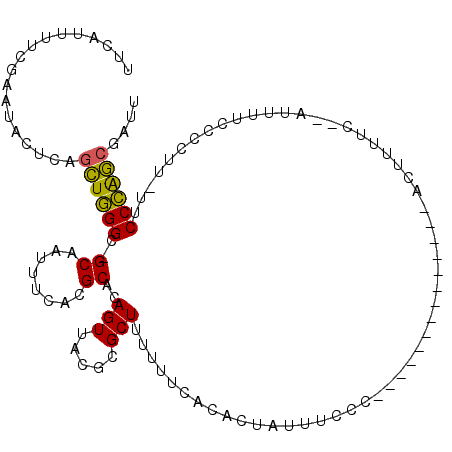
| Location | 20,094,118 – 20,094,219 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.13 |
| Mean single sequence MFE | -19.14 |
| Consensus MFE | -13.92 |
| Energy contribution | -13.70 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20094118 101 - 20766785 AAUCGCUGGGAAUAAGGGAAAAAUGGGAAAAGU--------------GGUAAAUAGCGUGAAAAAAGCGCGUAACUGUGCGUGAAAUUGC-GCCCAGCUGAGUAUUUGGAAAUGAU ..((((((((.....................(.--------------.((.(...((((.......)))).).))..)(((......)))-.)))))).))............... ( -20.80) >DroPse_CAF1 9124 110 - 1 AAUUGUUGGGAA-AAGCCGAAAA----AAAGGUUUGUUAAGACAUUAAAGGAAGGGUAUGAAAAAAGCGCGUAACUGUGCGUUAAAUUGC-GCCCAAGUUGAAAUUCGAAAAUUAA ...((((..((.-(((((.....----...))))).))..)))).........((((..((....((((((......))))))...))..-))))..................... ( -21.00) >DroSec_CAF1 8382 100 - 1 AAUCGCUGGGAA-AAGGGGAAAAUGGGAAAAGU--------------GGGAAAUAGCGUGAAAAAAGCGCGUAACUGUGCGUGAAAUUGC-GCCCAGCUGAGUAUUCGAAAAUGAA ..((((((((..-..................(.--------------.(..(...((((.......)))).)..)..)(((......)))-.)))))).))............... ( -20.20) >DroSim_CAF1 8362 100 - 1 AAUCGCUGGGAA-AAGGGGAAAAUGGGAAAAGU--------------GGGAAAUAGCGUGAAAAAAGCGCGUAACUGUGCGUGAAAUUGC-GCCCAGCUGAGUAUUCGAAAAUGAA ..((((((((..-..................(.--------------.(..(...((((.......)))).)..)..)(((......)))-.)))))).))............... ( -20.20) >DroEre_CAF1 8396 91 - 1 AACCGCCGGGAA-AA-UGG-------AA-AAGU--------------GGGAAAUAGUGUGAAAAAAGCGCGUAACUGUGCGUGAAAUUGC-GCCCAGCUGAGUAUUCGAAAAUGAA ..((((......-..-...-------..-..))--------------))(((...((((.......))))....(((.((((......))-)).))).......)))......... ( -15.46) >DroAna_CAF1 9017 97 - 1 AAUUGCUUGGGA-AAAGAGUAUAU----AAGCU--------------GGAAAAUUGUAUGAAAAAAGCGCGUAACUGUGCGUAAAAUUGCUGCCAAGCCAAAUAUUGUUAAUUGGC ....((((((..-......(((((----((..(--------------....).)))))))......(((((....)))))............)))))).................. ( -17.20) >consensus AAUCGCUGGGAA_AAGGGGAAAAU__GAAAAGU______________GGGAAAUAGCGUGAAAAAAGCGCGUAACUGUGCGUGAAAUUGC_GCCCAGCUGAGUAUUCGAAAAUGAA ....((((((........................................................(((((....)))))((......))..)))))).................. (-13.92 = -13.70 + -0.22)
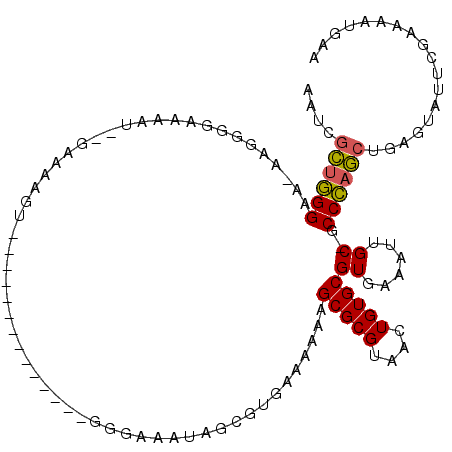
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:55 2006