
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
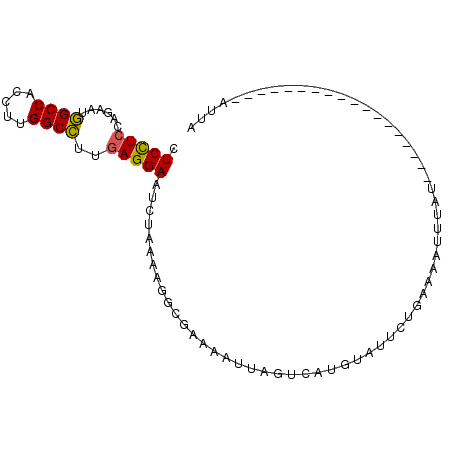
| Location | 20,086,508 – 20,086,602 |
| Length | 94 |
| Max. P | 0.856746 |

| Location | 20,086,508 – 20,086,602 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 64.20 |
| Mean single sequence MFE | -15.15 |
| Consensus MFE | -10.93 |
| Energy contribution | -10.55 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20086508 94 + 20766785 CUCUUCCAGAAUGGCCACCUUGGCCUUGAGGAAUCUAAAAGGCGAAAAUUAGUCAUGGUUUGUAAGAAUUCAUACUGUUUAGCUGAUCAU-AUUA ...((((..((.((((.....))))))..)))).......(((........)))((((((.((..((((.......)))).)).))))))-.... ( -17.20) >DroSec_CAF1 841 77 + 1 CUCUUCCAAAAUGGCCACCUUGGCCUUGAGGAAUCUAAAAGGCAAAAAUUAGUCAUGUAUUCUUAGAAUAUAU------------------AUUA ...((((..((.((((.....))))))..)))).......(((........)))((((((((...))))))))------------------.... ( -17.00) >DroEre_CAF1 836 77 + 1 CUCCUCCAGAAUGGCCACCUUGGCCUUGAGGAAUCUAAUAGACGAAAAUAAGUAUUGUAUUAUGGAAAUUUAU------------------AGUU .(((((......((((.....))))..))))).((((....((((.........))))....)))).......------------------.... ( -15.50) >DroAna_CAF1 766 75 + 1 CUCCUCCAGAAUAGCCACCUUGGCUUUAAGGAACCUGAAA------AAAGAGCCGUAU---CUGAAAGACAAU--CAAUU---------AAAUAA ......((((..((((.....))))....((...((....------..))..))...)---))).........--.....---------...... ( -10.90) >consensus CUCCUCCAGAAUGGCCACCUUGGCCUUGAGGAAUCUAAAAGGCGAAAAUUAGUCAUGUAUUCUGAAAAUUUAU__________________AUUA .(((((......((((.....))))..)))))............................................................... (-10.93 = -10.55 + -0.38)

| Location | 20,086,508 – 20,086,602 |
|---|---|
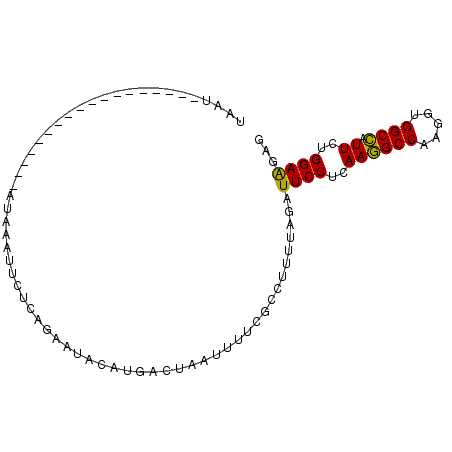
| Length | 94 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 64.20 |
| Mean single sequence MFE | -15.37 |
| Consensus MFE | -11.32 |
| Energy contribution | -10.70 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.71 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20086508 94 - 20766785 UAAU-AUGAUCAGCUAAACAGUAUGAAUUCUUACAAACCAUGACUAAUUUUCGCCUUUUAGAUUCCUCAAGGCCAAGGUGGCCAUUCUGGAAGAG ....-....((..(((((..((..(((((....((.....))...)))))..))..))))).((((..((((((.....)))).))..)))))). ( -14.80) >DroSec_CAF1 841 77 - 1 UAAU------------------AUAUAUUCUAAGAAUACAUGACUAAUUUUUGCCUUUUAGAUUCCUCAAGGCCAAGGUGGCCAUUUUGGAAGAG ....------------------.....((((((((.....(((..(((((.........)))))..))).((((.....)))).))))))))... ( -15.10) >DroEre_CAF1 836 77 - 1 AACU------------------AUAAAUUUCCAUAAUACAAUACUUAUUUUCGUCUAUUAGAUUCCUCAAGGCCAAGGUGGCCAUUCUGGAGGAG ....------------------.........................................(((((..((((.....))))......))))). ( -13.70) >DroAna_CAF1 766 75 - 1 UUAUUU---------AAUUG--AUUGUCUUUCAG---AUACGGCUCUUU------UUUCAGGUUCCUUAAAGCCAAGGUGGCUAUUCUGGAGGAG ......---------.....--....((((((((---(...((..(((.------....)))..))....((((.....))))..))))))))). ( -17.90) >consensus UAAU__________________AUAAAUUCUCAGAAUACAUGACUAAUUUUCGCCUUUUAGAUUCCUCAAGGCCAAGGUGGCCAUUCUGGAAGAG ..............................................................((((..((((((.....)))).))..))))... (-11.32 = -10.70 + -0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:53 2006