| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,080,657 – 20,080,767 |
| Length | 110 |
| Max. P | 0.686689 |

| Location | 20,080,657 – 20,080,767 |
|---|---|
| Length | 110 |
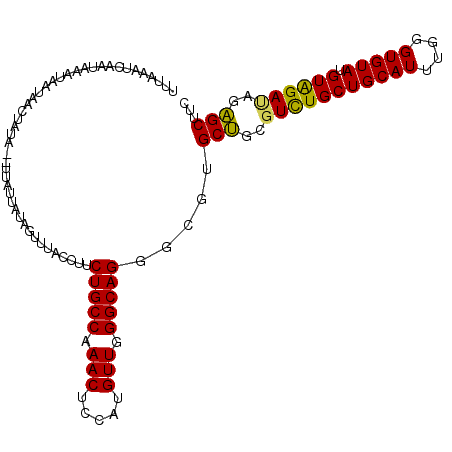
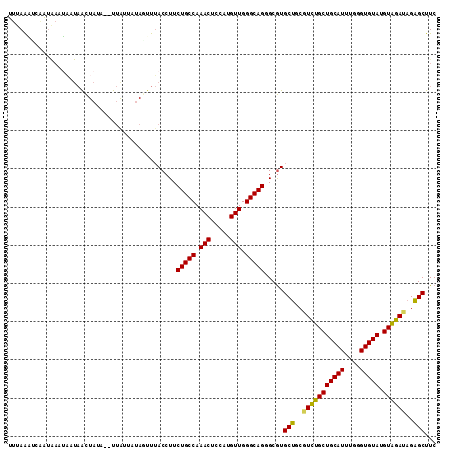
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 83.73 |
| Mean single sequence MFE | -28.83 |
| Consensus MFE | -24.93 |
| Energy contribution | -24.35 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20080657 110 - 20766785 UUUAAACCAAUAAAUAAUAACUAUU--UUAUUCUAGUUUACCUUCUGCCAAACUCCAUGUUGGGCAGGGCGUGCUUCGUCUGCUGCAUUUGGGUGUAUGUAGAUAGAGCUUC ..................(((((..--......))))).((.(((((((.(((.....))).))))))).))((((..((((((((((....))))).)))))..))))... ( -30.20) >DroVir_CAF1 3866 106 - 1 UUUAGUU-GAA---GUA--GUUACAAAUAAGGAGAACUAACCUUCUGCCAAACUCCAUGUUGGGCAGCGCAUGCGGCGUCUGCUGCAUUUGUGUGUAUGUGGAGAGCGCCUC ....((.-(((---(((--(((.(......)...)))))..)))).))....((((((((.(.(((.((.(((((((....))))))).))))).)))))))))........ ( -29.20) >DroSec_CAF1 4152 110 - 1 UUUAAAUCAAUAAAUAAUAACUAUA--UUACUUUAGUUUACCUUCUGCCAAACUCCAUGUUGGGCAGUGCGUGCUUCGUUUGCUGCAUUUGGGUGUAUGUAGAUAGAGCUUC ..................(((((..--......))))).((...(((((.(((.....))).)))))...))((((..((((((((((....))))).)))))..))))... ( -26.20) >DroSim_CAF1 4157 110 - 1 UUUAAAUCAAUAAAUAAUAACUAUA--UUACUUUAGUUUACCUUCUGCCAAACUCCAUGUUGGGCAGUGCGUGCUUCGUUUGCUGCAUUUGGGUGUAUGUAGAUAGAGCUUC ..................(((((..--......))))).((...(((((.(((.....))).)))))...))((((..((((((((((....))))).)))))..))))... ( -26.20) >DroEre_CAF1 4180 110 - 1 GUUAAAUCGUUGAAUAAUAACUAUU--UUGUUCUAUCUUACCUUCUGCCAAACUCCAUGUUGGGCAGGGCGUGCUGCGUCUGCUGCAUUUGGGUGUAUGUAGAUAGAGCUUC .........................--..((((((((((((((((((((.(((.....))).))))))..((((.((....)).))))..))))).....)))))))))... ( -31.10) >DroYak_CAF1 4116 110 - 1 GUUAAGUCUUUAAACUAUAACUAUU--UUAUUAUAUUUUACCUUCUGCCAAACUCCAUGUUGGGCAGGGCGUGCUGCGUCUGCUGCAUUUGGGUGUAUGUAGACAGAGCUUC (((((((......))).))))....--............((.(((((((.(((.....))).))))))).))(((..(((((((((((....))))).))))))..)))... ( -30.10) >consensus UUUAAAUCAAUAAAUAAUAACUAUA__UUAUUAUAGUUUACCUUCUGCCAAACUCCAUGUUGGGCAGGGCGUGCUGCGUCUGCUGCAUUUGGGUGUAUGUAGAUAGAGCUUC ............................................(((((.(((.....))).))))).....(((..(((((((((((....))))).))))))..)))... (-24.93 = -24.35 + -0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:50 2006