
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,075,455 – 20,075,557 |
| Length | 102 |
| Max. P | 0.582564 |

| Location | 20,075,455 – 20,075,557 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 83.50 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -26.51 |
| Energy contribution | -26.90 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
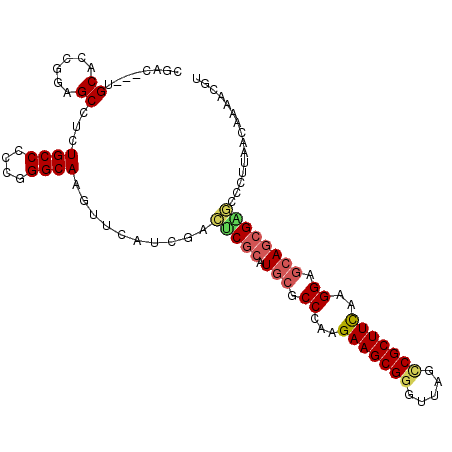
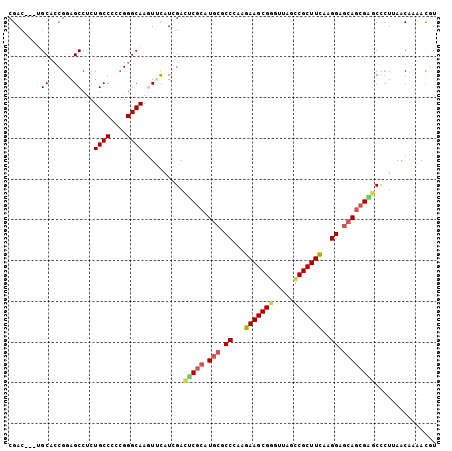
>2R_DroMel_CAF1 20075455 102 + 20766785 CGAC---UGCACCGGAGCCUCUGCCACCGGGCAAGUUCAUCGACUCGCAUGCGCCAAAGAAGCGGGUUAGCCGCUUCAAGGAGCAGCGAGCCCUUAACAAAACGU .(((---(((.((((.((....))..)))))).))))......(((((.(((.((...(((((((.....)))))))..)).))))))))............... ( -38.60) >DroVir_CAF1 2326 102 + 1 CGAC---AGCGCCGGAGCCACUGCCCGAGGGCAAAUAUAUAGAAACGCAUGCGCCCAAGAAGCGGAUCAGUCGCUUUAAGGAGCAACGCUCCAUGAACAAAAUAU ....---......(((((...((((....))))................(((..((..(((((((.....)))))))..)).)))..)))))............. ( -29.30) >DroGri_CAF1 2249 105 + 1 CGACAACGGCACCGGAGCCGCUGCCUGCGGGCAAAUACAUUGAUGCACAUACGCCAAAGAAGCGCGUUAGCCGCUUCAAGGAACAGCGUUCGCUGAGCAAGACAU ...(((((((......)))).((((....))))......))).(((.......((...(((((((....).))))))..))..((((....)))).)))...... ( -30.50) >DroSim_CAF1 2151 102 + 1 CGAC---UGCGCCGGAGCCUCUGCCCCCGGGCAAGUUCAUAGACUCGCAUGCGCCCAAGAAGCGGGUUAGCCGCUUCAAGGAGCAGCGAGCCCUUAACAAAACGU .(((---(((.((((.((....))..)))))).))))......(((((.(((..((..(((((((.....)))))))..)).))))))))............... ( -39.30) >DroEre_CAF1 2025 102 + 1 CGAC---UGCUGCGGAGCCUCUGCCUCCGGGCAAGUUCAUCGACUCGCAUGCGCCCAAGAAGCGGGUUAGCCGCUUCAAGGAGCAGCGGGCUCUUAACAAAACGU ....---......(((((((((((.(((((((.((((....)))).(....)))))..(((((((.....)))))))..))))))).)))))))........... ( -42.30) >DroYak_CAF1 2025 102 + 1 CGAC---UGCUCCGGAGCCUCUGCCCUCGGGCAAGUUCAUCGACUCGCAUGCGCCUAGGAAGCGGGUUAGCCGCUUCAAGGAGCAGCGAGCUCUUAACAAAACGU ....---.....((((((...((((....)))).))))...(((((((.(((.(((..(((((((.....))))))).))).)))))))).)).........)). ( -40.20) >consensus CGAC___UGCACCGGAGCCUCUGCCCCCGGGCAAGUUCAUCGACUCGCAUGCGCCCAAGAAGCGGGUUAGCCGCUUCAAGGAGCAGCGAGCCCUUAACAAAACGU ........((......))...((((....))))..........(((((.(((.((...(((((((.....)))))))..)).))))))))............... (-26.51 = -26.90 + 0.39)

| Location | 20,075,455 – 20,075,557 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.50 |
| Mean single sequence MFE | -42.45 |
| Consensus MFE | -29.04 |
| Energy contribution | -29.10 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
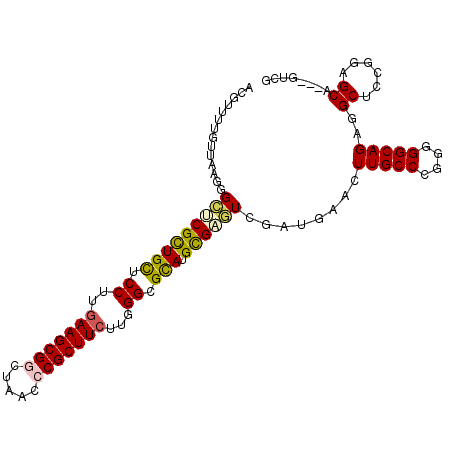
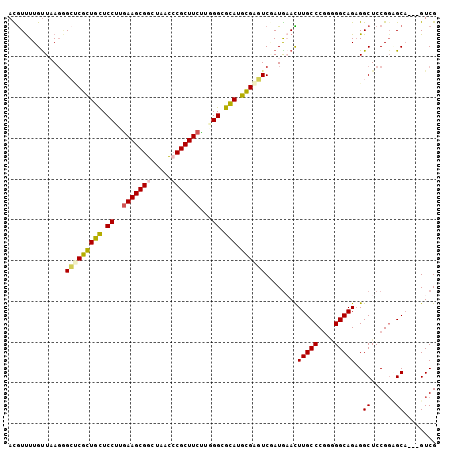
>2R_DroMel_CAF1 20075455 102 - 20766785 ACGUUUUGUUAAGGGCUCGCUGCUCCUUGAAGCGGCUAACCCGCUUCUUUGGCGCAUGCGAGUCGAUGAACUUGCCCGGUGGCAGAGGCUCCGGUGCA---GUCG .......(((...((((((((((.((..(((((((.....)))))))...)).))).)))))))....)))((((((((.(((....))))))).)))---)... ( -45.90) >DroVir_CAF1 2326 102 - 1 AUAUUUUGUUCAUGGAGCGUUGCUCCUUAAAGCGACUGAUCCGCUUCUUGGGCGCAUGCGUUUCUAUAUAUUUGCCCUCGGGCAGUGGCUCCGGCGCU---GUCG .......((...(((((((((((.(((..(((((.......)))))...))).))).))............(((((....)))))..))))))..)).---.... ( -31.40) >DroGri_CAF1 2249 105 - 1 AUGUCUUGCUCAGCGAACGCUGUUCCUUGAAGCGGCUAACGCGCUUCUUUGGCGUAUGUGCAUCAAUGUAUUUGCCCGCAGGCAGCGGCUCCGGUGCCGUUGUCG ......(((.((((....))))......((((((.(....)))))))...((.(((.(((((....))))).))))))))(((((((((......))))))))). ( -41.80) >DroSim_CAF1 2151 102 - 1 ACGUUUUGUUAAGGGCUCGCUGCUCCUUGAAGCGGCUAACCCGCUUCUUGGGCGCAUGCGAGUCUAUGAACUUGCCCGGGGGCAGAGGCUCCGGCGCA---GUCG .......(((..((((((((((((((..(((((((.....)))))))..))).))).))))))))...)))((((((((((.(...).)))))).)))---)... ( -49.80) >DroEre_CAF1 2025 102 - 1 ACGUUUUGUUAAGAGCCCGCUGCUCCUUGAAGCGGCUAACCCGCUUCUUGGGCGCAUGCGAGUCGAUGAACUUGCCCGGAGGCAGAGGCUCCGCAGCA---GUCG .((..(((((..(((((..(((((((..(((((((.....)))))))..(((((....)((((......))))))))))).)))).)))))...))))---).)) ( -41.10) >DroYak_CAF1 2025 102 - 1 ACGUUUUGUUAAGAGCUCGCUGCUCCUUGAAGCGGCUAACCCGCUUCCUAGGCGCAUGCGAGUCGAUGAACUUGCCCGAGGGCAGAGGCUCCGGAGCA---GUCG ..((((......((.((((((((.(((.(((((((.....)))))))..))).))).)))))))...))))(((((((.((((....))))))).)))---)... ( -44.70) >consensus ACGUUUUGUUAAGGGCUCGCUGCUCCUUGAAGCGGCUAACCCGCUUCUUGGGCGCAUGCGAGUCGAUGAACUUGCCCGGGGGCAGAGGCUCCGGAGCA___GUCG ..............(((((((((.((..(((((((.....)))))))...)).))).))))))........(((((....)))))..((......))........ (-29.04 = -29.10 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:46 2006