
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,067,871 – 20,067,973 |
| Length | 102 |
| Max. P | 0.530359 |

| Location | 20,067,871 – 20,067,973 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -19.27 |
| Energy contribution | -19.38 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20067871 102 - 20766785 CAUUCCACUGUACGCCAACGAGGAGAAAACUAUCUACAACAUGGUGGUGGAGGUGCCACGUUGGACCAACGCGAAAAUGGAGGUCGGUC-UGCCUGUCCCUA---------A ...((((.....(((((......(((......)))......)))))(((..(((.((.....)))))..))).....))))((.(((..-...))).))...---------. ( -24.90) >DroVir_CAF1 30716 100 - 1 CAUUCCAUUGUACGCCAACGAGGAGAAGACCAUCUACAACAUGGUCGUCGAGGUGCCACGUUGGACGAACGCCAAAAUGGAGGUAAAU------AG------CAUAAUUAUC ..(((((((((.((((((((.((.((.((((((.......)))))).))......)).)))))).)).)).....)))))))......------..------.......... ( -27.50) >DroPse_CAF1 4680 91 - 1 CAUUCCAUUGUACGCCAAUGAGGAGAAGACCAUCUACAACAUGGUGGUGGAGGUUCCACGUUGGACCAACGCCAAGAUGGAGGUAAGC------------AA---------A ..(((((((....((((.((..((((......))).)..))))))((((..(((((......)))))..))))..)))))))......------------..---------. ( -26.00) >DroWil_CAF1 833 100 - 1 CAUUCCAUUGUAUGCCAAUGAGGAGAAGACCGUCUACAAUAUGGUUGUUGAGGUGCCCCGUUGGACAAACGCCAAAAUGGAGGUAAGCA-AACAUGUCUCA----------- .....(((((.....)))))..((((.((((((.......))))))(((....(((((((((((.(....))))..)))).))))....-)))...)))).----------- ( -24.50) >DroAna_CAF1 4894 103 - 1 CAUUCCCCUCUACGCCAAUGAGGAUAAGUCCAUCUACAACAUGGUGGUGGAGGUGCCACGUUGGACCAACGCCAAGAUGGAGGUGGGCAAUGAUUGAUCCAU---------A ((.((.((((.........))))....((((((((....(((..(((((..(((.((.....)))))..)))))..))).))))))))...)).))......---------. ( -35.40) >DroPer_CAF1 8991 91 - 1 CAUUCCAUUGUACGCCAAUGAGGAGAAGACCAUCUACAACAUGGUGGUGGAGGUUCCACGUUGGACCAACGCCAAGAUGGAGGUAAGC------------AA---------A ..(((((((....((((.((..((((......))).)..))))))((((..(((((......)))))..))))..)))))))......------------..---------. ( -26.00) >consensus CAUUCCAUUGUACGCCAAUGAGGAGAAGACCAUCUACAACAUGGUGGUGGAGGUGCCACGUUGGACCAACGCCAAAAUGGAGGUAAGC______UG____AA_________A ..(((((((...((((((((.((.....(((.(((((.((...)).)))))))).)).)))))).....))....))))))).............................. (-19.27 = -19.38 + 0.11)

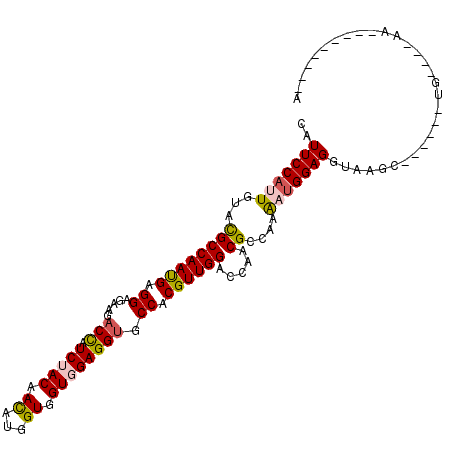
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:42 2006