
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,065,408 – 20,065,509 |
| Length | 101 |
| Max. P | 0.993482 |

| Location | 20,065,408 – 20,065,509 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.43 |
| Mean single sequence MFE | -18.94 |
| Consensus MFE | -8.24 |
| Energy contribution | -8.08 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
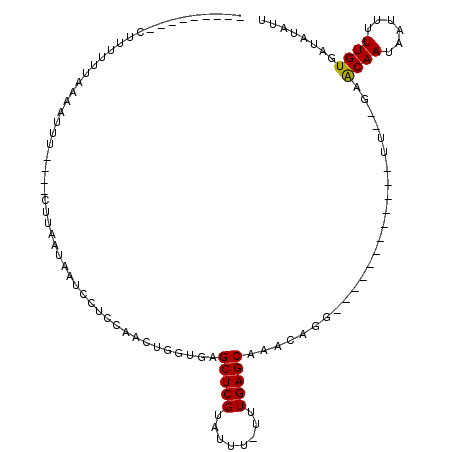
>2R_DroMel_CAF1 20065408 101 + 20766785 ---------UUUUUGUUAAAUUC----CUCAAUAAUCCUCGAACUGUUGAGCUCGUAUUU-UUUGAGCAAACAAACAGAACAAGAUUUUGAGAACAAUAAUUUUUGUGAUAUAUU ---------..............----(((((.((((......(((((..(((((.....-..))))).....))))).....))))))))).((((......))))........ ( -17.20) >DroSec_CAF1 2427 84 + 1 ---------CUUUUUUAAAAUU-------UAAUAAUCCUCCAACUGGUGAGCUCGUAUUU-UUUGAGCAAACAGU------------UU--GAACAAUAAUUUUUGUGAUAUAUU ---------.............-------.....(((.((.(((((....(((((.....-..)))))...))))------------).--))((((......)))))))..... ( -13.50) >DroSim_CAF1 2439 87 + 1 ---------CUUUUUUAAAAUUU----CUUAAUAAUCCUUCAACUGGUGAGCUCGUAUUU-UUUGAGCAAACAGG------------UU--GAACAAUAAUUUUUGUGAUAUAUU ---------..............----.......(((..((((((.((..(((((.....-..)))))..)).))------------))--))((((......)))))))..... ( -16.40) >DroEre_CAF1 2506 96 + 1 UUCGAAACUCGUUUUUCAAGUUU----CUGAGUAAUUUUCUAACUGUUGUGCUCGUAUUU-UUUGAGCAUAUCGG------------UU--GAACAAUAAUUUUUGUGCUAUAUU ...((((((.(.....).)))))----)..(((.......((((((.((((((((.....-..)))))))).)))------------))--).((((......)))))))..... ( -23.20) >DroYak_CAF1 2436 101 + 1 UUCUAAACUCUUUUUUCAAAUUACAUACUUAGUAAUUUUCUAACUGGUGUGCUCGUAUUUUUUUGAGCAAACCGG------------UU--GAGCAAUAAUUUUUGUGAAAUAUU .............(((((((((((.......)))))))..((((((((.((((((........)))))).)))))------------))--).((((......)))))))).... ( -24.40) >consensus _________CUUUUUUAAAAUUU____CUUAAUAAUCCUCCAACUGGUGAGCUCGUAUUU_UUUGAGCAAACAGG____________UU__GAACAAUAAUUUUUGUGAUAUAUU ..................................................(((((........))))).........................((((......))))........ ( -8.24 = -8.08 + -0.16)

| Location | 20,065,408 – 20,065,509 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.43 |
| Mean single sequence MFE | -17.30 |
| Consensus MFE | -4.30 |
| Energy contribution | -4.30 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.25 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
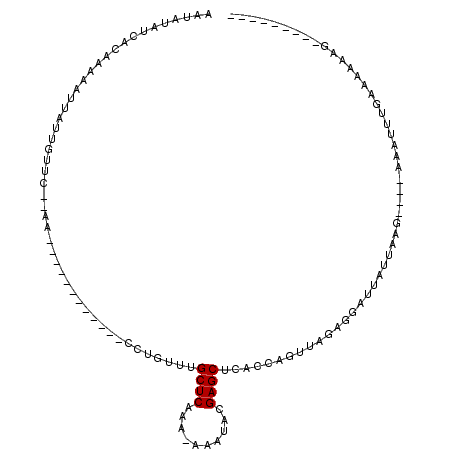
>2R_DroMel_CAF1 20065408 101 - 20766785 AAUAUAUCACAAAAAUUAUUGUUCUCAAAAUCUUGUUCUGUUUGUUUGCUCAAA-AAAUACGAGCUCAACAGUUCGAGGAUUAUUGAG----GAAUUUAACAAAAA--------- ...............(((...((((((((((((((..(((((.(...((((...-......)))).))))))..).)))))).)))))----))...)))......--------- ( -21.50) >DroSec_CAF1 2427 84 - 1 AAUAUAUCACAAAAAUUAUUGUUC--AA------------ACUGUUUGCUCAAA-AAAUACGAGCUCACCAGUUGGAGGAUUAUUA-------AAUUUUAAAAAAG--------- .............(((.(((.(((--((------------.(((...((((...-......))))....)))))))).))).))).-------.............--------- ( -12.90) >DroSim_CAF1 2439 87 - 1 AAUAUAUCACAAAAAUUAUUGUUC--AA------------CCUGUUUGCUCAAA-AAAUACGAGCUCACCAGUUGAAGGAUUAUUAAG----AAAUUUUAAAAAAG--------- .............(((.(((.(((--((------------(..((..((((...-......))))..))..)))))).))).)))...----..............--------- ( -14.80) >DroEre_CAF1 2506 96 - 1 AAUAUAGCACAAAAAUUAUUGUUC--AA------------CCGAUAUGCUCAAA-AAAUACGAGCACAACAGUUAGAAAAUUACUCAG----AAACUUGAAAAACGAGUUUCGAA ....((((........(((((...--..------------.)))))(((((...-......))))).....))))............(----(((((((.....))))))))... ( -17.30) >DroYak_CAF1 2436 101 - 1 AAUAUUUCACAAAAAUUAUUGCUC--AA------------CCGGUUUGCUCAAAAAAAUACGAGCACACCAGUUAGAAAAUUACUAAGUAUGUAAUUUGAAAAAAGAGUUUAGAA ....................((((--((------------(.(((.(((((..........))))).))).)))...(((((((.......))))))).......))))...... ( -20.00) >consensus AAUAUAUCACAAAAAUUAUUGUUC__AA____________CCUGUUUGCUCAAA_AAAUACGAGCUCACCAGUUAGAGGAUUAUUAAG____AAAUUUGAAAAAAG_________ ...............................................((((..........)))).................................................. ( -4.30 = -4.30 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:40 2006