
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,063,676 – 20,063,776 |
| Length | 100 |
| Max. P | 0.626758 |

| Location | 20,063,676 – 20,063,776 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 85.11 |
| Mean single sequence MFE | -20.28 |
| Consensus MFE | -16.22 |
| Energy contribution | -17.13 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20063676 100 + 20766785 GCAGCUGCAGCACUAUCUGAAAGUGAUCAACGGAUCACCCUAUCCGUUUCACAAGCCCAUCUACCACUAGACUCCUAAC-UUCGACCACUUUCGU------UUCCAC ...((....)).......(((((((.(((((((((......)))))))...........((((....))))........-...)).)))))))..------...... ( -19.90) >DroSec_CAF1 723 88 + 1 GCAGCUGCAGCACUAUCUGAAAGUGAUCAACGGAUCGCCCUAUCCGUUUCACAAGCCCGUCUACCACUAGACUCCUAAA-UUCGGCCAC------------------ ...((....)).....(((((.((((..(((((((......)))))))))))......(((((....))))).......-)))))....------------------ ( -20.50) >DroSim_CAF1 713 100 + 1 GCAGCUGCAGCACUAUCUGAAAGUGAUCAACGGAUCGCCCUAUCCGUUUCACAAGCCCGUCUACCACUAGACUCCUAGC-UUCGGCCACUUUCGU------UUCCAC ...((((.(((...........((((..(((((((......)))))))))))......(((((....))))).....))-).)))).........------...... ( -25.40) >DroEre_CAF1 738 105 + 1 GCAGCUGCAGCACUAUCUGAAAGUGAUCAACGGAUCGCCCUAUCCGUUUCACAAGCCCGUCUACCACUAGACUCCUUAC-UUCGACCACUUCCGUUUC-GUUUCCAC ...((....)).......((((((((..(((((((......)))))))))))......(((((....))))).......-..............))))-........ ( -19.00) >DroYak_CAF1 710 106 + 1 GCAGCUGCAGCACUAUCUGAAAGUGAUCAACGGAUCGCCCUAUCCGUUUCACAAGCCCGUCUACCACUAGACUCCUUAC-UUCGACCACUUUCGUUUCUACUUCCAC ...((....)).......(((((((.(((((((((......)))))))..........(((((....))))).......-...)).))))))).............. ( -23.30) >DroAna_CAF1 694 102 + 1 GCAGCUCCAGCACUAUCUAAAAGUGAUCAAUGGAUCGCCAUAUCCAUUCCACAAGCCCAAAUACCAUUAGAUUUCUUACCUACUGUCACUUUCGUU-----UUCCGC ((.((....))........(((((((..(((((((......)))))))................((.(((.........))).)))))))))....-----....)) ( -13.60) >consensus GCAGCUGCAGCACUAUCUGAAAGUGAUCAACGGAUCGCCCUAUCCGUUUCACAAGCCCGUCUACCACUAGACUCCUAAC_UUCGACCACUUUCGU______UUCCAC ...((....)).......(((((((.(((((((((......)))))))..........(((((....)))))...........)).))))))).............. (-16.22 = -17.13 + 0.92)

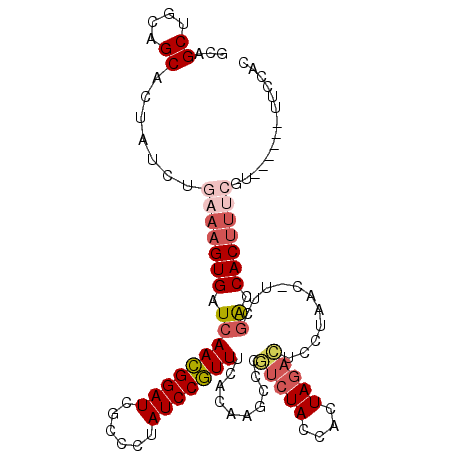
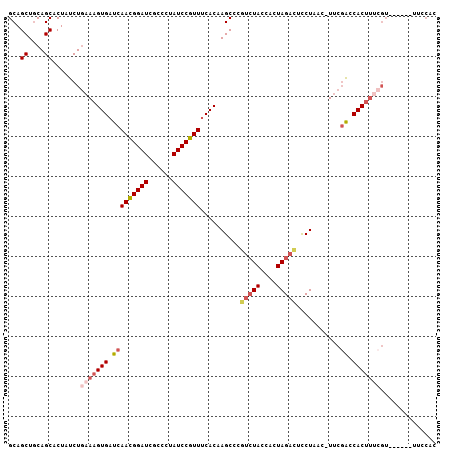
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:36 2006