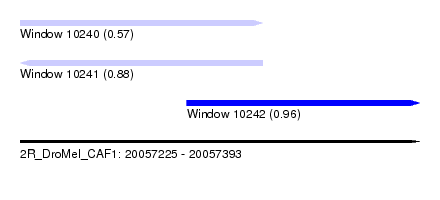
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,057,225 – 20,057,393 |
| Length | 168 |
| Max. P | 0.959224 |

| Location | 20,057,225 – 20,057,327 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 74.43 |
| Mean single sequence MFE | -21.64 |
| Consensus MFE | -12.01 |
| Energy contribution | -12.35 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20057225 102 + 20766785 UUCCGACUCAAACUUUGACUCAAAGCU--GGCUGCAAAGAGCCUCGCUGCAAAACA---ACAACACAUACAUAUGCAUACGAAAGCCGCACUGAGUCGAGUAUGAUA .......(((.((((.((((((..((.--((((((.....)).(((.((((.....---..............))))..))).))))))..)))))))))).))).. ( -24.91) >DroEre_CAF1 17313 80 + 1 UUCCGACUCAAACUUUGGCUCAAAGC----------------CUCGCUGCAAAACA---GAAAC--------ACAUAUACGAAACCCGCACUGAGUCGAGCAUGAUA ...(((((((......(((.....))----------------)...(((.....))---)....--------...................)))))))......... ( -15.80) >DroYak_CAF1 17334 96 + 1 UUCCGACUCAAACUUUGGCUCAAAGCUCUGG-----------CUGGCUGCAAAACAACAGCAACACAUAUACACAUGUACGAAAGCCGCACUGAGUCGAGCAUGGUA ...(((((((......(((.....)))..((-----------((.((((........))))...((((......)))).....))))....)))))))......... ( -24.20) >consensus UUCCGACUCAAACUUUGGCUCAAAGCU__GG___________CUCGCUGCAAAACA___GCAACACAUA_A_ACAUAUACGAAAGCCGCACUGAGUCGAGCAUGAUA ...(((((((..(((((...)))))....................((.((..................................)).))..)))))))......... (-12.01 = -12.35 + 0.33)



| Location | 20,057,225 – 20,057,327 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 74.43 |
| Mean single sequence MFE | -28.00 |
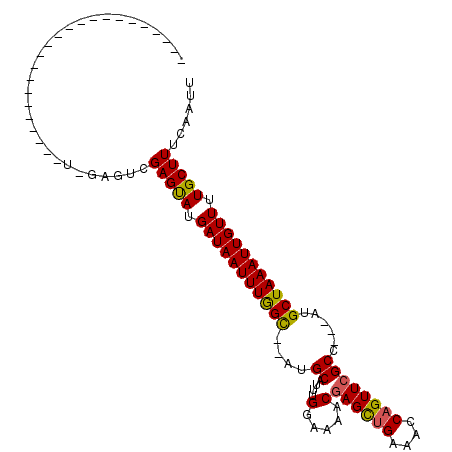
| Consensus MFE | -19.97 |
| Energy contribution | -19.87 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20057225 102 - 20766785 UAUCAUACUCGACUCAGUGCGGCUUUCGUAUGCAUAUGUAUGUGUUGU---UGUUUUGCAGCGAGGCUCUUUGCAGCC--AGCUUUGAGUCAAAGUUUGAGUCGGAA ..(((.(((.(((((((.((((((.....((((....))))((.((((---((.....)))))).)).......))))--.)).)))))))..))).)))....... ( -31.40) >DroEre_CAF1 17313 80 - 1 UAUCAUGCUCGACUCAGUGCGGGUUUCGUAUAUGU--------GUUUC---UGUUUUGCAGCGAG----------------GCUUUGAGCCAAAGUUUGAGUCGGAA ........(((((((((.((.((((.((.....((--------.((.(---((.....))).)).----------------))..))))))...))))))))))).. ( -22.10) >DroYak_CAF1 17334 96 - 1 UACCAUGCUCGACUCAGUGCGGCUUUCGUACAUGUGUAUAUGUGUUGCUGUUGUUUUGCAGCCAG-----------CCAGAGCUUUGAGCCAAAGUUUGAGUCGGAA ........(((((((.....((((....((((((....))))))..(((((......))))).))-----------)).((((((((...))))))))))))))).. ( -30.50) >consensus UAUCAUGCUCGACUCAGUGCGGCUUUCGUAUAUGU_U_UAUGUGUUGC___UGUUUUGCAGCGAG___________CC__AGCUUUGAGCCAAAGUUUGAGUCGGAA ........(((((((((((((.....)))))............(((((.........)))))...................((((((...)))))).)))))))).. (-19.97 = -19.87 + -0.11)

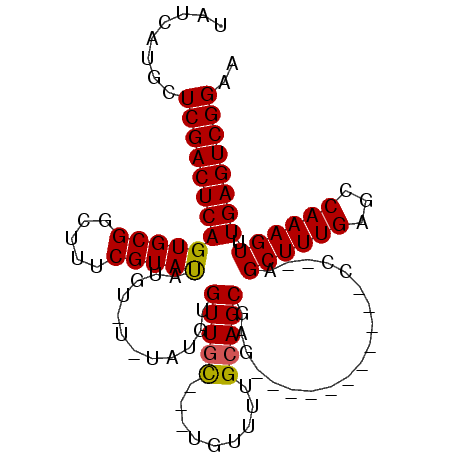
| Location | 20,057,295 – 20,057,393 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.56 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -20.80 |
| Energy contribution | -20.58 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20057295 98 + 20766785 CAUAC-------GAAAGCCGCACU-GAGUCGAGUAUGAUAAUUUGGC--AUGCAUUUGGAAAACGAGCUGAAACCAGUUCGCC---AUGCUAAAUUGUUUUGCUUUCAAUU .....-------((((((...(((-......)))..(((((((((((--(((...((....))(((((((....))))))).)---)))))))))))))..)))))).... ( -30.50) >DroSec_CAF1 16900 82 + 1 -----------------------U-GAGUCGAGUAUGAUAAUUUGGC--AUGCAUUUGGAAAACGAGCUGAAACCAGUUCGCC---AUGCUAAAUUGUUUUGCUUUCAAUU -----------------------(-((...(((((.(((((((((((--(((...((....))(((((((....))))))).)---))))))))))))).))))))))... ( -28.50) >DroSim_CAF1 17382 82 + 1 -----------------------U-GAGUCGAGUAUGAUAAUUUGGC--AUGCAUUUGGAAAACGAGCUGAAACCAGUUCGCC---AUGCUAAAUUGUUUUGCUUUCAAUU -----------------------(-((...(((((.(((((((((((--(((...((....))(((((((....))))))).)---))))))))))))).))))))))... ( -28.50) >DroEre_CAF1 17361 98 + 1 UAUAC-------GAAACCCGCACU-GAGUCGAGCAUGAUAAUUUGGC--AUGCAUUUGGAAAACGAGCUGAAACCAGUUCGCC---AUGCUAAAUUGUUUUGCUUUCAAUU .....-------...........(-((...(((((.(((((((((((--(((...((....))(((((((....))))))).)---))))))))))))).))))))))... ( -30.60) >DroWil_CAF1 17463 109 + 1 UCUACAUCCUCCGUUUGCCGU--UGCCAUCGAGUAUGAUAAUUUAGUUGAAGCAUUUGGAAAACGAGUCGAAACCAGUUCGCCAUCAUGCUAAAUUGUUUUGCUUUCAAUU .....................--.......(((((.((((((((((((((......(((..(((..((....))..)))..)))))).))))))))))).)))))...... ( -19.20) >DroAna_CAF1 18913 84 + 1 ---------------------ACU-GCAUCGAGUAUGAUAAUUUGGC--GUGCGUUUGGAAAACAAGCUGAAACCAGUUCGCC---AUGCUAAAUUGUUUUGCUUUCAAUU ---------------------...-.....(((((.(((((((((((--(((((((((.....))))).(((.....)))).)---))))))))))))).)))))...... ( -25.10) >consensus _______________________U_GAGUCGAGUAUGAUAAUUUGGC__AUGCAUUUGGAAAACGAGCUGAAACCAGUUCGCC___AUGCUAAAUUGUUUUGCUUUCAAUU ..............................(((((.(((((((((((....((....(.....)((((((....))))))))......))))))))))).)))))...... (-20.80 = -20.58 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:31 2006