
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,054,075 – 20,054,215 |
| Length | 140 |
| Max. P | 0.973558 |

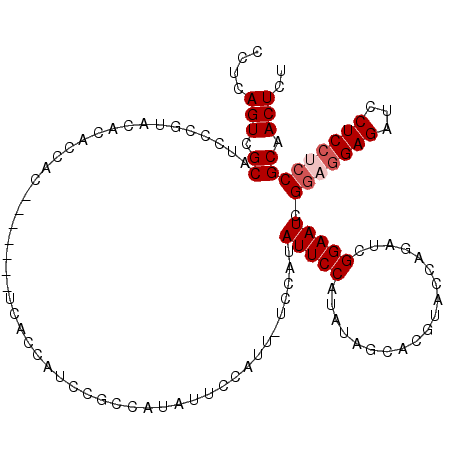
| Location | 20,054,075 – 20,054,177 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 85.61 |
| Mean single sequence MFE | -18.82 |
| Consensus MFE | -12.71 |
| Energy contribution | -13.71 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20054075 102 + 20766785 CCUCAGUCGCAUCCCGUACACACCAC-------UCACCAUCCGCCAUAU-------UCCAUAUUCCAUAUAGCACGUACCAGAUCGGAAUCGGAGGAGAUCCUCCGCCGCAACUCU ....(((.((................-------......(((((.((((-------..........)))).)).(......)...)))..((((((....))))))..)).))).. ( -17.90) >DroSec_CAF1 13856 103 + 1 CCUCAGUCGCAUCCCGUACACACCAC-------U------CUGCCAUAUUCCAUUCUCCAUAUUCCAUAUAGCACGUACCAGAUCGGAAUCGGAGGAGAUCCUCCUCCGCAACUCU ....(((.((...(((........((-------.------.(((.((((.................)))).))).)).......)))....(((((((...))))))))).))).. ( -19.89) >DroSim_CAF1 14223 109 + 1 CCUCAGUCGCAUCCCGUACACACCAC-------UCACCAUCCGCCAUAUUCCAUUCUCCAUAUUCCAUAUAGCACGUACCAGAUCGGAAUCGGAGGAGAUCCUCCUCCGCAACUCU ....(((.((................-------..............(((((..(((.......................)))..))))).(((((((...))))))))).))).. ( -19.20) >DroEre_CAF1 14122 112 + 1 CCUCAGUCGCAUCCCGUAUACACCACUCACCAUUCGCCAUUCGCCAUAUUCCCAU-UCCAUAUUCCAUCUAGCCCGUAGCAGAUCGGAAUCG---GAGAUCCUCCUCCGCAACUCU ....(((.((.............................................-.....(((((((((.((.....)))))).))))).(---(((......)))))).))).. ( -18.30) >consensus CCUCAGUCGCAUCCCGUACACACCAC_______UCACCAUCCGCCAUAUUCCAUU_UCCAUAUUCCAUAUAGCACGUACCAGAUCGGAAUCGGAGGAGAUCCUCCUCCGCAACUCU ....(((.((...................................................(((((...................))))).(((((((...))))))))).))).. (-12.71 = -13.71 + 1.00)


| Location | 20,054,075 – 20,054,177 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 85.61 |
| Mean single sequence MFE | -35.97 |
| Consensus MFE | -26.70 |
| Energy contribution | -28.26 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20054075 102 - 20766785 AGAGUUGCGGCGGAGGAUCUCCUCCGAUUCCGAUCUGGUACGUGCUAUAUGGAAUAUGGA-------AUAUGGCGGAUGGUGA-------GUGGUGUGUACGGGAUGCGACUGAGG ..((((((((((((((....))))))...)).((((.(((((..((((........((..-------......))........-------))))..))))).)))))))))).... ( -36.39) >DroSec_CAF1 13856 103 - 1 AGAGUUGCGGAGGAGGAUCUCCUCCGAUUCCGAUCUGGUACGUGCUAUAUGGAAUAUGGAGAAUGGAAUAUGGCAG------A-------GUGGUGUGUACGGGAUGCGACUGAGG ..(((((((((((((...))))))).......((((.(((((..((((.((..((((..........))))..)).------.-------))))..))))).)))))))))).... ( -39.60) >DroSim_CAF1 14223 109 - 1 AGAGUUGCGGAGGAGGAUCUCCUCCGAUUCCGAUCUGGUACGUGCUAUAUGGAAUAUGGAGAAUGGAAUAUGGCGGAUGGUGA-------GUGGUGUGUACGGGAUGCGACUGAGG ..(((((((((((((...))))))).......((((.(((((..(((((((...(((.....)))...))).((.....))..-------))))..))))).)))))))))).... ( -36.50) >DroEre_CAF1 14122 112 - 1 AGAGUUGCGGAGGAGGAUCUC---CGAUUCCGAUCUGCUACGGGCUAGAUGGAAUAUGGA-AUGGGAAUAUGGCGAAUGGCGAAUGGUGAGUGGUGUAUACGGGAUGCGACUGAGG ..((((((((((......)))---).((((((((..(((((..((((..((..((.((..-(((....)))..)).))..))..))))..)))))..)).)))))))))))).... ( -31.40) >consensus AGAGUUGCGGAGGAGGAUCUCCUCCGAUUCCGAUCUGGUACGUGCUAUAUGGAAUAUGGA_AAUGGAAUAUGGCGGAUGGUGA_______GUGGUGUGUACGGGAUGCGACUGAGG ..((((((((((((((....)))))...))).((((.(((((..((((.((..((((..........))))..))...............))))..))))).)))))))))).... (-26.70 = -28.26 + 1.56)

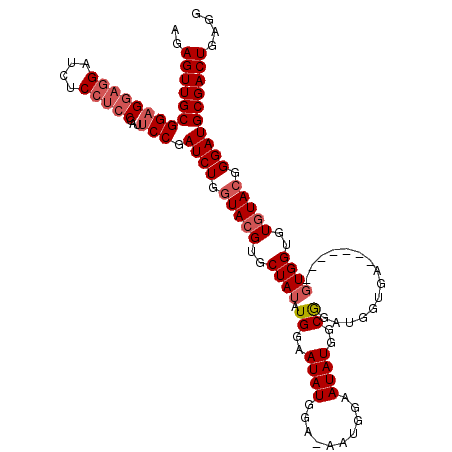
| Location | 20,054,104 – 20,054,215 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 90.38 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -23.64 |
| Energy contribution | -24.45 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20054104 111 + 20766785 CCAUCCGCCAUAU-------UCCAUAUUCCAUAUAGCACGUACCAGAUCGGAAUCGGAGGAGAUCCUCCGCCGCAACUCUGGCUAAUUGAAAAGCAAUUGUGGCGCACUCAACUAGCA .....((((((..-------......................(((((.(((...((((((....)))))))))....)))))..(((((.....)))))))))))............. ( -29.50) >DroSec_CAF1 13883 114 + 1 ----CUGCCAUAUUCCAUUCUCCAUAUUCCAUAUAGCACGUACCAGAUCGGAAUCGGAGGAGAUCCUCCUCCGCAACUCUGGCUAAUUGAAAAGCAAUUGUGGCGCACUCAACUAGCC ----.(((...(((((..(((.......................)))..))))).(((((((...)))))))))).....(((((.((((...((.......))....)))).))))) ( -28.10) >DroSim_CAF1 14252 118 + 1 CCAUCCGCCAUAUUCCAUUCUCCAUAUUCCAUAUAGCACGUACCAGAUCGGAAUCGGAGGAGAUCCUCCUCCGCAACUCUGGCUAAUUGAAAAGCAAUUGUGGCGCACUCAACUAGCC .....((((((...............................(((((.((((...(((((....)))))))))....)))))..(((((.....)))))))))))............. ( -29.00) >DroEre_CAF1 14158 114 + 1 CCAUUCGCCAUAUUCCCAU-UCCAUAUUCCAUCUAGCCCGUAGCAGAUCGGAAUCG---GAGAUCCUCCUCCGCAACUCUGGCUAAUUGAAAAGCAAUUGUGGCGCACUCAACUAGCC .....((((((....(((.-.....(((((((((.((.....)))))).)))))((---(((......)))))......)))..(((((.....)))))))))))............. ( -27.00) >consensus CCAUCCGCCAUAUUCCAUU_UCCAUAUUCCAUAUAGCACGUACCAGAUCGGAAUCGGAGGAGAUCCUCCUCCGCAACUCUGGCUAAUUGAAAAGCAAUUGUGGCGCACUCAACUAGCC .....(((((((...............................((((.((((...(((((....)))))))))....))))(((........)))...)))))))............. (-23.64 = -24.45 + 0.81)


| Location | 20,054,104 – 20,054,215 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 90.38 |
| Mean single sequence MFE | -38.80 |
| Consensus MFE | -31.33 |
| Energy contribution | -32.58 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20054104 111 - 20766785 UGCUAGUUGAGUGCGCCACAAUUGCUUUUCAAUUAGCCAGAGUUGCGGCGGAGGAUCUCCUCCGAUUCCGAUCUGGUACGUGCUAUAUGGAAUAUGGA-------AUAUGGCGGAUGG .((((((((((.(((.......)))..))))))))))((((....(((((((((....))))))...))).)))).....((((((((..........-------))))))))..... ( -38.40) >DroSec_CAF1 13883 114 - 1 GGCUAGUUGAGUGCGCCACAAUUGCUUUUCAAUUAGCCAGAGUUGCGGAGGAGGAUCUCCUCCGAUUCCGAUCUGGUACGUGCUAUAUGGAAUAUGGAGAAUGGAAUAUGGCAG---- (((((((((((.(((.......)))..)))))))))))(((....(((((((((....)))))...)))).)))......((((((((....(((.....)))..)))))))).---- ( -40.40) >DroSim_CAF1 14252 118 - 1 GGCUAGUUGAGUGCGCCACAAUUGCUUUUCAAUUAGCCAGAGUUGCGGAGGAGGAUCUCCUCCGAUUCCGAUCUGGUACGUGCUAUAUGGAAUAUGGAGAAUGGAAUAUGGCGGAUGG (((((((((((.(((.......)))..))))))))))).......((((((((...))))))))..((((........(((.((((((...))))))...)))........))))... ( -40.09) >DroEre_CAF1 14158 114 - 1 GGCUAGUUGAGUGCGCCACAAUUGCUUUUCAAUUAGCCAGAGUUGCGGAGGAGGAUCUC---CGAUUCCGAUCUGCUACGGGCUAGAUGGAAUAUGGA-AUGGGAAUAUGGCGAAUGG (((((((((((.(((.......)))..)))))))))))....((((((((......)))---).(((((.((((((.....)).))))))))).....-...........)))).... ( -36.30) >consensus GGCUAGUUGAGUGCGCCACAAUUGCUUUUCAAUUAGCCAGAGUUGCGGAGGAGGAUCUCCUCCGAUUCCGAUCUGGUACGUGCUAUAUGGAAUAUGGA_AAUGGAAUAUGGCGGAUGG .((((((((((.(((.......)))..))))))))))((((....(((((((((....)))))...)))).))))......(((((((.................)))))))...... (-31.33 = -32.58 + 1.25)

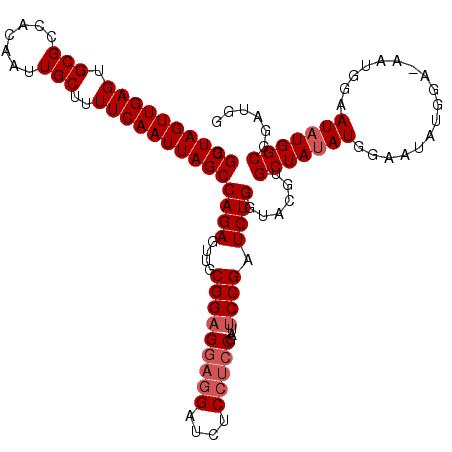

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:28 2006