| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,053,601 – 20,053,754 |
| Length | 153 |
| Max. P | 0.956438 |

| Location | 20,053,601 – 20,053,719 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.30 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -18.29 |
| Energy contribution | -18.22 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
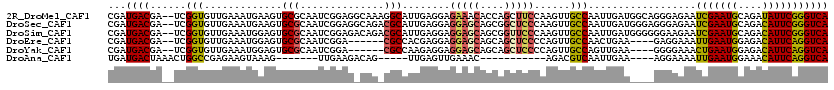
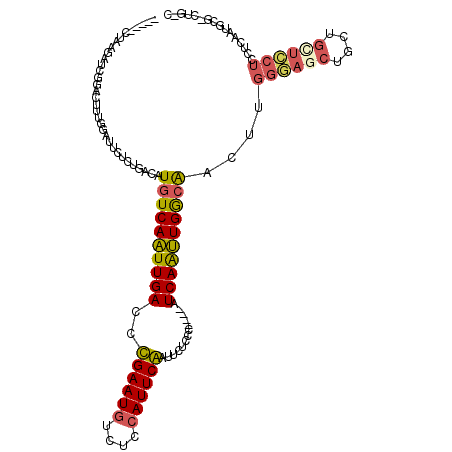
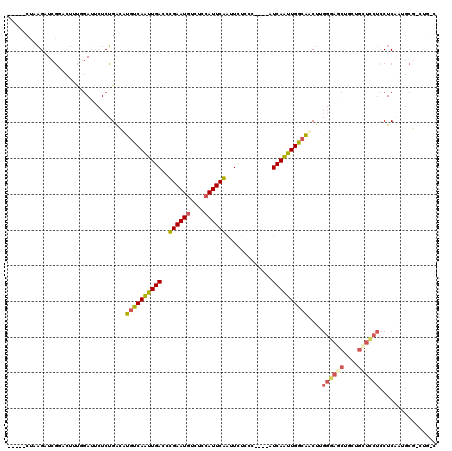
>2R_DroMel_CAF1 20053601 118 + 20766785 CGAUGACGA--UCGGUGUUGAAAUGAAGUGCGCAAUCGGAGGCAAAGGCAUUGAGGAGAAACACCAGCUUCCAAGUUGCCAAUUGAUGGCAGGGAGAAUCGAAUGCAGAUAUUCGGGUCA .......((--((((((((.......(((((((........))....))))).......))))))..(((((....(((((.....))))))))))...((((((....)))))))))). ( -32.44) >DroSec_CAF1 13368 118 + 1 CGAUGACGA--UCGGUGUUGAAAUGAAGUGCGCAAUCGGAGGCAGACGCAUUGAGGAGGAGCAGCGGCUCCCAAGUUGCCAAUUGAUGGGAGGGAGAAUCGAAUGCAGACAUUCGGGUCA ...((((..--(((.((((.......((((((...((.......)))))))).......)))).)))(((((....(.(((.....))).))))))..(((((((....))))))))))) ( -31.24) >DroSim_CAF1 13718 118 + 1 CGAUGACGA--UCGGUGUUGAAAUGGAGUGCGCAAUCGGAGACAGACGCAUUGAGGAGGAGCAGCGGUUCCCAAGUUGCCAAUUGAUGGGGGGAAGAAUCGAAUGCAGACAUUCGGGUCA ...((((((--(((.((((....(..((((((...(((....).))))))))..)....)))).)))))(((..(((.......)))..)))......(((((((....))))))))))) ( -31.20) >DroEre_CAF1 13625 108 + 1 CGAUGACGA--UCGGUGUUGAAAUGGAGUGCGCAAUCGGA------CGCCACGAGGAGGAGCAGCAGCUCCCCAGUUGCCAACUGAA----GAGGAAAUUGAAUGGAGACAUUCAGGUCA ((((..((.--((.((......)).))...))..))))((------(((.((..((.(((((....))))))).)).))........----.......(((((((....)))))))))). ( -35.90) >DroYak_CAF1 13683 108 + 1 CGAUGACGA--UCGGUGUUGAAAUGGAGUGCGCAAUCGGA------CGCCAAGAGGAGGAGCAGCAGCUCCCCAGUUGCCAGUUGAA----GGGGAAACUGAAUGGAGACAUUCAGGUCA .(((..(((--(.(((.......(((.((.((....)).)------).)))...((.(((((....)))))))....))).))))..----.......(((((((....)))))))))). ( -38.70) >DroAna_CAF1 15691 93 + 1 UGAUGACUAAACUGGCCGAGAAGUAAAG-------UUGAAGACAG-----UUGAGUUGAAAC-----------AGACGUCAAUUGAA----AGGAAAAUUGAAUGGAAACAUUCAGGUCA .............((((...........-------.......(((-----(((((((.....-----------.))).)))))))..----........((((((....)))))))))). ( -23.50) >consensus CGAUGACGA__UCGGUGUUGAAAUGAAGUGCGCAAUCGGAG_CAG_CGCAUUGAGGAGGAGCAGCAGCUCCCAAGUUGCCAAUUGAA____GGGAAAAUCGAAUGCAGACAUUCAGGUCA ...((((......(((.............(((..............)))........(((((....)))))......)))..................(((((((....))))))))))) (-18.29 = -18.22 + -0.07)
| Location | 20,053,639 – 20,053,754 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.48 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -18.30 |
| Energy contribution | -18.02 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
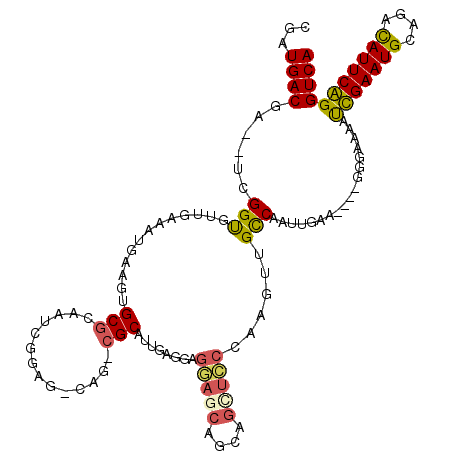
>2R_DroMel_CAF1 20053639 115 + 20766785 GGCAAAGGCAUUGAGGAGAAACACCAGCUUCCAAGUUGCCAAUUGAUGGCAGGGAGAAUCGAAUGCAGAUAUUCGGGUCAAUUGACAACGCAGAGAAUCAAUAGUACGAUCUCAG----- .((....(((((((((.......))..(((((....(((((.....))))))))))..)).)))))..........(((....)))...)).((((.((........))))))..----- ( -26.70) >DroSec_CAF1 13406 115 + 1 GGCAGACGCAUUGAGGAGGAGCAGCGGCUCCCAAGUUGCCAAUUGAUGGGAGGGAGAAUCGAAUGCAGACAUUCGGGUCAAUUGACAUGUCAGAGAAUCCAAAGUACGAACUCAG----- .((....))..(((((.(((((....))))))...((.(((.....))).))(((((..((((((....))))))..))..((((....))))....)))..........)))).----- ( -29.10) >DroSim_CAF1 13756 115 + 1 GACAGACGCAUUGAGGAGGAGCAGCGGUUCCCAAGUUGCCAAUUGAUGGGGGGAAGAAUCGAAUGCAGACAUUCGGGUCAAUUGACAUGUCAGAGAAUCCAAAGUACGAACUCAG----- ((((...(((.....(.(((((....))))))....)))((((((((...........(((((((....)))))))))))))))...)))).(((..((........)).)))..----- ( -26.40) >DroEre_CAF1 13663 105 + 1 ------CGCCACGAGGAGGAGCAGCAGCUCCCCAGUUGCCAACUGAA----GAGGAAAUUGAAUGGAGACAUUCAGGUCAAUUGACAAGCCAGAGCAUCUGUAGUCCGAUCUUAG----- ------.((.((..((.(((((....))))))).)).))......((----(((((..(((((((....)))))))(((....)))....(((.....)))...)))..))))..----- ( -32.10) >DroYak_CAF1 13721 105 + 1 ------CGCCAAGAGGAGGAGCAGCAGCUCCCCAGUUGCCAGUUGAA----GGGGAAACUGAAUGGAGACAUUCAGGUCAACUGAAAAGUCAGAGAAUCCGCAGUCCGAUCUUAG----- ------....((((((.(((((....)))))))..(((((((((((.----.......(((((((....))))))).)))))))........(......))))).....))))..----- ( -34.40) >DroAna_CAF1 15724 100 + 1 GACAG-----UUGAGUUGAAAC-----------AGACGUCAAUUGAA----AGGAAAAUUGAAUGGAAACAUUCAGGUCAAUUGACAUGUCUGAAGCUGGCAAAUCAAAUCAUUGAUUGG ..(((-----(..(.((((..(-----------(((((((((((((.----.......(((((((....))))))).)))))))))..)))))..(....)...))))....)..)))). ( -32.50) >consensus G_CAG_CGCAUUGAGGAGGAGCAGCAGCUCCCAAGUUGCCAAUUGAA____GGGAAAAUCGAAUGCAGACAUUCAGGUCAAUUGACAAGUCAGAGAAUCCAAAGUACGAUCUCAG_____ ............(((..(((((....)))))........(((((((............(((((((....))))))).)))))))..........................)))....... (-18.30 = -18.02 + -0.28)
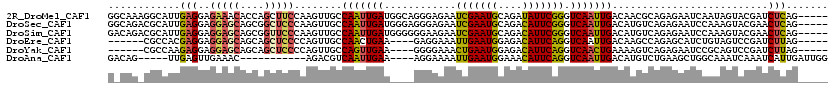
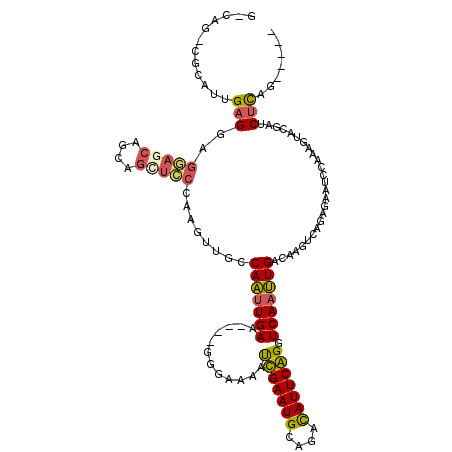
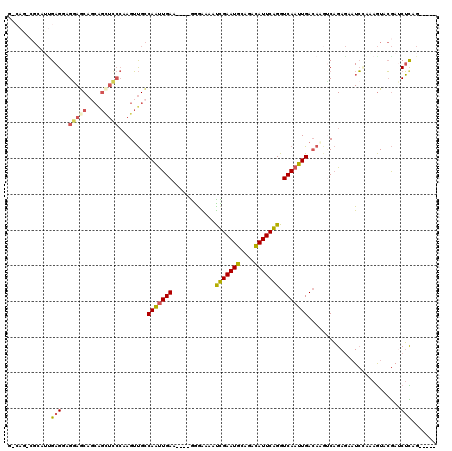
| Location | 20,053,639 – 20,053,754 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.48 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -17.61 |
| Energy contribution | -17.61 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20053639 115 - 20766785 -----CUGAGAUCGUACUAUUGAUUCUCUGCGUUGUCAAUUGACCCGAAUAUCUGCAUUCGAUUCUCCCUGCCAUCAAUUGGCAACUUGGAAGCUGGUGUUUCUCCUCAAUGCCUUUGCC -----..(((((((......))).)))).((((((((((((((..(((((......)))))...(.....)...))))))))))))..((((((....)))))).......))....... ( -26.80) >DroSec_CAF1 13406 115 - 1 -----CUGAGUUCGUACUUUGGAUUCUCUGACAUGUCAAUUGACCCGAAUGUCUGCAUUCGAUUCUCCCUCCCAUCAAUUGGCAACUUGGGAGCCGCUGCUCCUCCUCAAUGCGUCUGCC -----..(((((((.....)))))))...(((.((((((((((..((((((....)))))).............))))))))))....((((((....)))))).........))).... ( -31.86) >DroSim_CAF1 13756 115 - 1 -----CUGAGUUCGUACUUUGGAUUCUCUGACAUGUCAAUUGACCCGAAUGUCUGCAUUCGAUUCUUCCCCCCAUCAAUUGGCAACUUGGGAACCGCUGCUCCUCCUCAAUGCGUCUGUC -----..(((((((.....)))))))...(((.((((((((((...(((((....)))))((....))......))))))))))....((((........)))).........))).... ( -27.00) >DroEre_CAF1 13663 105 - 1 -----CUAAGAUCGGACUACAGAUGCUCUGGCUUGUCAAUUGACCUGAAUGUCUCCAUUCAAUUUCCUC----UUCAGUUGGCAACUGGGGAGCUGCUGCUCCUCCUCGUGGCG------ -----.........(.((((.........((.(((((((((((..((((((....))))))........----.)))))))))))))(((((((....)))))))...))))).------ ( -30.40) >DroYak_CAF1 13721 105 - 1 -----CUAAGAUCGGACUGCGGAUUCUCUGACUUUUCAGUUGACCUGAAUGUCUCCAUUCAGUUUCCCC----UUCAACUGGCAACUGGGGAGCUGCUGCUCCUCCUCUUGGCG------ -----((((((..((((.((((.((((((...((.((((((((.(((((((....))))))).......----.)))))))).))..)))))))))).).)))...))))))..------ ( -32.70) >DroAna_CAF1 15724 100 - 1 CCAAUCAAUGAUUUGAUUUGCCAGCUUCAGACAUGUCAAUUGACCUGAAUGUUUCCAUUCAAUUUUCCU----UUCAAUUGACGUCU-----------GUUUCAACUCAA-----CUGUC ........(((.((((...........(((((..(((((((((..((((((....))))))........----.)))))))))))))-----------)..)))).))).-----..... ( -20.82) >consensus _____CUAAGAUCGGACUUUGGAUUCUCUGACAUGUCAAUUGACCCGAAUGUCUCCAUUCAAUUCUCCC____AUCAAUUGGCAACUUGGGAGCUGCUGCUCCUCCUCAAUGCG_CUG_C .................................((((((((((..((((((....)))))).............))))))))))....((((((....))))))................ (-17.61 = -17.61 + 0.00)
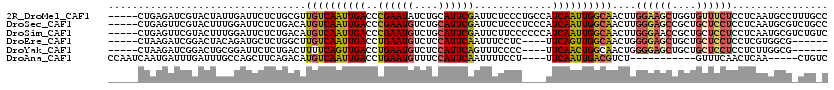
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:24 2006