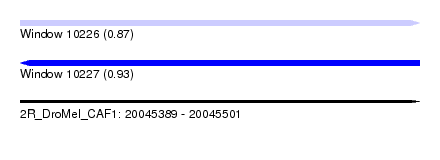
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,045,389 – 20,045,501 |
| Length | 112 |
| Max. P | 0.929390 |

| Location | 20,045,389 – 20,045,501 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 89.14 |
| Mean single sequence MFE | -38.04 |
| Consensus MFE | -27.63 |
| Energy contribution | -27.75 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.869828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20045389 112 + 20766785 AGAACCAUGUCCCAACCAAAACGACACUAUCAUUUUGCUUGGGAUUUUAUGGCCACACAAACCCAUC-------CACGGAUCGAUCGAUUGAUCGUUUGCGGUCGAUUGGGGCUAAGUG ....(((((((((((.(((((.((.....)).))))).)))))))...))))...(((...((((((-------....)))....((((((((((....)))))))))))))....))) ( -34.80) >DroSec_CAF1 5276 119 + 1 AGAACCAUGUCCCAACCAAAACGACACUAUCAUUUUGCGUGGGAUUUUAUGGCCACAUAAGCCCAUCCACGAUCCACGGAUCGAUCGAUUGAUCGUUUGCGGUCGAUUGGGGCUAAGUG ....((((((((((..(((((.((.....)).)))))..))))))...)))).(((...(((((.....(((((....)))))..((((((((((....)))))))))))))))..))) ( -44.90) >DroSim_CAF1 5341 119 + 1 AGAACCAUGUCCCAACCAAAACGACACUUUCAUUUUGCGUGGGAUUUUAUGGCCACAUAAGCCCAUCCACGAUCCACGGAUCGAUCGAUUGAUCGUUUGCGGUCGAUUGGGGCUAAGUG ....((((((((((..(((((.((.....)).)))))..))))))...)))).(((...(((((.....(((((....)))))..((((((((((....)))))))))))))))..))) ( -45.00) >DroEre_CAF1 5404 112 + 1 AGAACCAUGUCCCAACCAAUAAGACACUAGUAUUUCGCGUGGGAUUUUAUGGCCACAUGAACCCAUC-------CACGGAUCGAUCGAUUGAUCGCUUGCGGUCGAUUGAGGCUAAGUG ........................(((((((...((.((((((..((((((....))))))....))-------))))))....(((((((((((....))))))))))).))).)))) ( -31.50) >DroYak_CAF1 5403 112 + 1 AGAACCAUGUCCCCACCAAUAUGGGACUAUUAUUUCACGUGGGACUUUAUGGCCACAUAAACCCAUC-------CACGGAUCGAUCGAUUGAUCGCUUGCGGUCGAUUGGGGCUGAGUG ((..((..(((((.........)))))..........((((((..((((((....))))))....))-------))))(((((((((...(....)...)))))))))))..))..... ( -34.00) >consensus AGAACCAUGUCCCAACCAAAACGACACUAUCAUUUUGCGUGGGAUUUUAUGGCCACAUAAACCCAUC_______CACGGAUCGAUCGAUUGAUCGUUUGCGGUCGAUUGGGGCUAAGUG ...((...(((((........(((..(...........(((((..((((((....)))))))))))...........)..)))...(((((((((....))))))))))))))...)). (-27.63 = -27.75 + 0.12)

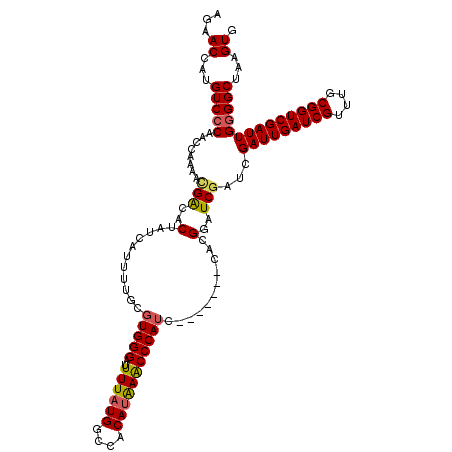

| Location | 20,045,389 – 20,045,501 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.14 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -29.33 |
| Energy contribution | -30.01 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20045389 112 - 20766785 CACUUAGCCCCAAUCGACCGCAAACGAUCAAUCGAUCGAUCCGUG-------GAUGGGUUUGUGUGGCCAUAAAAUCCCAAGCAAAAUGAUAGUGUCGUUUUGGUUGGGACAUGGUUCU (((..(((((..(((.((.(....(((((....)))))..).)).-------))))))))...)))(((((....((((((.(((((((((...))))))))).)))))).)))))... ( -40.40) >DroSec_CAF1 5276 119 - 1 CACUUAGCCCCAAUCGACCGCAAACGAUCAAUCGAUCGAUCCGUGGAUCGUGGAUGGGCUUAUGUGGCCAUAAAAUCCCACGCAAAAUGAUAGUGUCGUUUUGGUUGGGACAUGGUUCU (((..(((((....(((((((...(((((....)))))....)))).))).....)))))...)))(((((....((((((.(((((((((...)))))))))).))))).)))))... ( -44.30) >DroSim_CAF1 5341 119 - 1 CACUUAGCCCCAAUCGACCGCAAACGAUCAAUCGAUCGAUCCGUGGAUCGUGGAUGGGCUUAUGUGGCCAUAAAAUCCCACGCAAAAUGAAAGUGUCGUUUUGGUUGGGACAUGGUUCU (((..(((((....(((((((...(((((....)))))....)))).))).....)))))...)))(((((....((((((.((((((((.....))))))))).))))).)))))... ( -43.80) >DroEre_CAF1 5404 112 - 1 CACUUAGCCUCAAUCGACCGCAAGCGAUCAAUCGAUCGAUCCGUG-------GAUGGGUUCAUGUGGCCAUAAAAUCCCACGCGAAAUACUAGUGUCUUAUUGGUUGGGACAUGGUUCU .....((((........((((...(((((....)))))....)))-------).(((.(((.(((((..........))))).)))...)))((((((((.....)))))))))))).. ( -29.40) >DroYak_CAF1 5403 112 - 1 CACUCAGCCCCAAUCGACCGCAAGCGAUCAAUCGAUCGAUCCGUG-------GAUGGGUUUAUGUGGCCAUAAAGUCCCACGUGAAAUAAUAGUCCCAUAUUGGUGGGGACAUGGUUCU ...(((.(((((.((((((((...(((((....)))))....)))-------)((((((((((((((..........)))))))))........))))).)))))))))...))).... ( -36.50) >consensus CACUUAGCCCCAAUCGACCGCAAACGAUCAAUCGAUCGAUCCGUG_______GAUGGGUUUAUGUGGCCAUAAAAUCCCACGCAAAAUGAUAGUGUCGUUUUGGUUGGGACAUGGUUCU (((..(((((........(((...(((((....)))))....)))..........)))))...)))(((((....(((((..((((((((.....))))))))..))))).)))))... (-29.33 = -30.01 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:17 2006