
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,027,211 – 20,027,325 |
| Length | 114 |
| Max. P | 0.700274 |

| Location | 20,027,211 – 20,027,325 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.52 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -12.42 |
| Energy contribution | -13.71 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20027211 114 + 20766785 AACAACGCGUAUCCACAGGAUACUUUCAACUCACAGAUGCUUGCACUUGCCUGCUUGCAAACCGAAAGCGAAG---AUGCAGUGAAGUUGCUUUAUAGAAGUCUUAAGGAUAUGCGA .....(((((((((..(((((.....((((((((...(((..(((......)))..)))........((....---..)).))).)))))((....))..)))))..))))))))). ( -33.10) >DroSec_CAF1 44228 113 + 1 AAGAACGCGUAUCCACAAGAUACUUUCAACUCGCAGAUACUUGCACUUGCCUGCUUGCAGAACCAAAGCGAAG---AUGCAGUAAAAUCGA-UUAUAGAAGUCUUGAGGAUAUGCGA .....(((((((((.(((((..((((....(((....((((.((((((...(((((.........))))))))---.)))))))....)))-.....))))))))).))))))))). ( -33.30) >DroSim_CAF1 45095 114 + 1 AAGAACGCGUAUCCACAAGAUACUUUCAACUCGCAGAUACUUGCACUUGCCUGCUUGCAGAACCAAAGCGAAA---AGGCAGCGAAAUCGCUUUAUAGAAGUCUUGAGGAUAUGCGA .....(((((((((.((((((...(((.....((((....))))...(((((..((((.........))))..---)))))(((....)))......))))))))).))))))))). ( -40.50) >DroEre_CAF1 50313 114 + 1 AGCAACGCGCAUCCGAAAGGUAUUUUCAACACACGGAUGCUGGCACUGGCCUGCUUGCAGAACCAAAGCGGAG---AUGCAGUGAAGUCGCUGUAUGGCAACCUGGAUGGCAUGCGA .(((..((.((((((((((...))))).......((.((((.((..(((.(((....)))..)))..))....---((((((((....)))))))))))).)).))))))).))).. ( -38.10) >DroYak_CAF1 49781 114 + 1 CGCAGCGCGUAUCUGAAAGAUACUUUCAACUCUGAGAUGCUAGCACUGGCGUGCUUGCAGAACAACAGCGAAG---AUGCACUGAAGUCGCUGUACAGCAGUCUUGAGGGCAUCCGA .....((((((((.....)))))(((((....)))))(((.(((((....))))).)))........)))..(---((((.((.(((.((((....))).).))).)).)))))... ( -36.20) >DroPer_CAF1 46364 114 + 1 CAUUCCCUUUAUCUGACGAAGAUAUUCAACUCGGAGCUUCUGGCCGUGGUUUGCGA---AAACCGCAGUGAAGGCGACACCAUAAAGGCUCUUUAUAGAAAUCUACAGGACAUUCGC .........(((((.....))))).......((((..(((((((((((((((....---))))))).(((..(....)..)))...)))......(((....))))))))..)))). ( -26.30) >consensus AACAACGCGUAUCCAAAAGAUACUUUCAACUCGCAGAUGCUUGCACUGGCCUGCUUGCAGAACCAAAGCGAAG___AUGCAGUGAAGUCGCUUUAUAGAAGUCUUGAGGACAUGCGA .....(((((.(((..(((((.....................(((......)))..........(((((((................)))))))......)))))..))).))))). (-12.42 = -13.71 + 1.28)

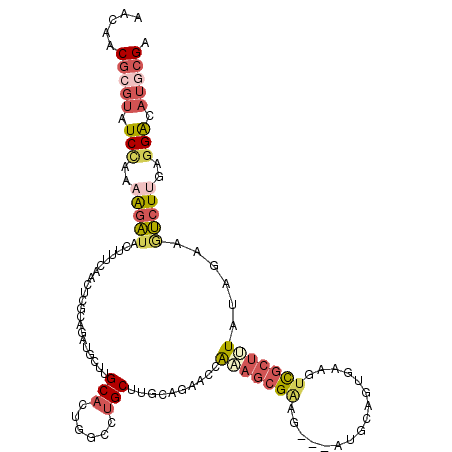
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:06 2006