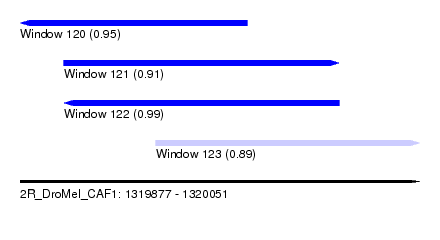
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,319,877 – 1,320,051 |
| Length | 174 |
| Max. P | 0.992079 |

| Location | 1,319,877 – 1,319,976 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 86.87 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -22.04 |
| Energy contribution | -23.85 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1319877 99 - 20766785 UGGCCGACUGCCCGGACAAACGAGGCAGAUAAAAAACGAUUGCAGCGAUACAAGCUGUAAAUAAAGAGGACCCUGGGAUCCGAAUCGGGGACACACAGC ...((..(((((((......)).)))))...........(((((((.......))))))).......)).((((((........))))))......... ( -30.70) >DroSec_CAF1 38415 98 - 1 UGGCCGACUGCCC-GACAAACGAGGCAGAUAAAAAACGAUUGCAGCGAUACGAGCUGUAAAUAAAGAGGACCCUGGGAUCCGAAUCGGGGACACACAGC ...((..((((((-(.....)).)))))...........(((((((.......))))))).......)).((((((........))))))......... ( -29.20) >DroSim_CAF1 17493 99 - 1 UGGCCGACUGCCCGGACAAACGAGGCAGAUAAAAAACGAUUGCAGCGAUACGAGCUGUAAAUAAAGAGGACCCUGGGAUCCGAAUCGGGGACACACAGC ...((..(((((((......)).)))))...........(((((((.......))))))).......)).((((((........))))))......... ( -30.70) >DroYak_CAF1 5949 83 - 1 UGGCCGACUGUCCGGACAAACGAGGCAGAUAAAAAACUAUUGCAGCGAUACGAGCUGUAAAUAAAGGGAAACCUGGUAUCAA----------------C ..(((..(((((((......)).)))))...........(((((((.......)))))))....(((....)))))).....----------------. ( -26.40) >consensus UGGCCGACUGCCCGGACAAACGAGGCAGAUAAAAAACGAUUGCAGCGAUACGAGCUGUAAAUAAAGAGGACCCUGGGAUCCGAAUCGGGGACACACAGC ...((..(((((((......)).)))))...........(((((((.......))))))).......)).((((((........))))))......... (-22.04 = -23.85 + 1.81)

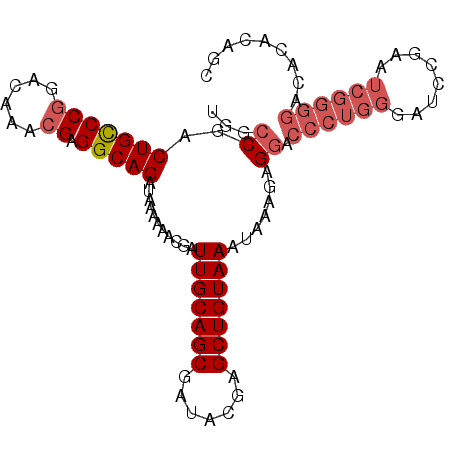
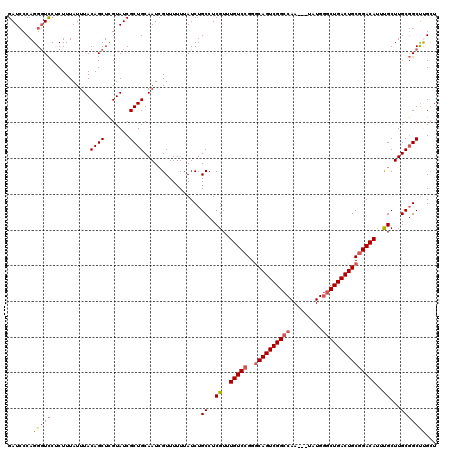
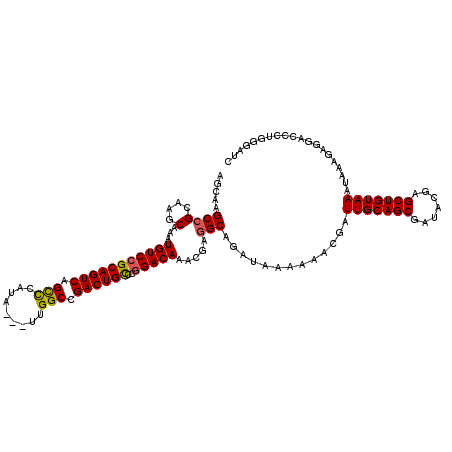
| Location | 1,319,896 – 1,320,016 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.98 |
| Mean single sequence MFE | -37.78 |
| Consensus MFE | -32.36 |
| Energy contribution | -32.92 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1319896 120 + 20766785 GAUCCCAGGGUCCUCUUUAUUUACAGCUUGUAUCGCUGCAAUCGUUUUUUAUCUGCCUCGUUUGUCCGGGCAGUCGGCCAAUAAUAUGGGCUGACUGCGGACAUUUGCUUGCGGCCUGCU .....((((..............((((.......))))..............((((..((..(((((..((((((((((.........)))))))))))))))..))...)))))))).. ( -39.60) >DroSec_CAF1 38434 116 + 1 GAUCCCAGGGUCCUCUUUAUUUACAGCUCGUAUCGCUGCAAUCGUUUUUUAUCUGCCUCGUUUGUC-GGGCAGUCGGCCAA---UAUGGGCUGACUGCGGACAUUUGCUUGCGGCUCGCU ((((....))))................((..((((.((((..((((.....(((.(......).)-))((((((((((..---....))))))))))))))..))))..))))..)).. ( -37.10) >DroSim_CAF1 17512 117 + 1 GAUCCCAGGGUCCUCUUUAUUUACAGCUCGUAUCGCUGCAAUCGUUUUUUAUCUGCCUCGUUUGUCCGGGCAGUCGGCCAA---UAUGGGCUGACUGCGGACAUUUGCUUGCGGCUCGCU .......(((((...........((((.......))))................((..((..(((((..((((((((((..---....)))))))))))))))..))...)))))))... ( -39.20) >DroEre_CAF1 19923 117 + 1 GAUCCCAGGGUCCGCUUUAUUUACAGCUCGUAUCGCUGCAAUCGUUUUUUAUCUGCCUCGUUUGUCCGGGCAGUCGGCCAA---UAUGGGCUGACUGCGGACACUCGCUUGCGGCUUGGU ....(((((..((((........((((.......))))....................((..(((((..((((((((((..---....)))))))))))))))..))...))))))))). ( -43.60) >DroYak_CAF1 5952 117 + 1 GAUACCAGGUUUCCCUUUAUUUACAGCUCGUAUCGCUGCAAUAGUUUUUUAUCUGCCUCGUUUGUCCGGACAGUCGGCCAA---UAUGAACUGACUGCGGACAUUCGCUUGCCGCUUGCU (((((..((((.............)))).)))))((.(((((((........)))...((..((((((..(((((((....---......)))))))))))))..)).)))).))..... ( -29.42) >consensus GAUCCCAGGGUCCUCUUUAUUUACAGCUCGUAUCGCUGCAAUCGUUUUUUAUCUGCCUCGUUUGUCCGGGCAGUCGGCCAA___UAUGGGCUGACUGCGGACAUUUGCUUGCGGCUUGCU .......(((((...........((((.......))))................((..((..(((((..((((((((((.........)))))))))))))))..))...)))))))... (-32.36 = -32.92 + 0.56)

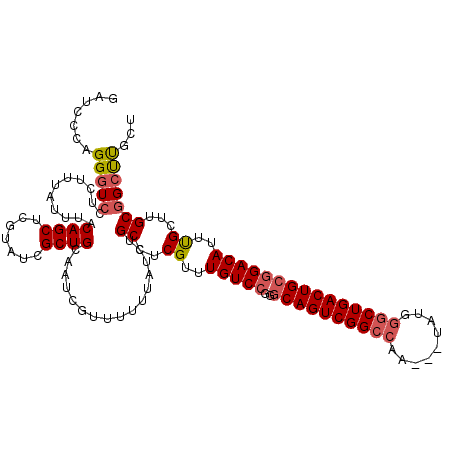
| Location | 1,319,896 – 1,320,016 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.98 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -31.18 |
| Energy contribution | -31.10 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1319896 120 - 20766785 AGCAGGCCGCAAGCAAAUGUCCGCAGUCAGCCCAUAUUAUUGGCCGACUGCCCGGACAAACGAGGCAGAUAAAAAACGAUUGCAGCGAUACAAGCUGUAAAUAAAGAGGACCCUGGGAUC ..((((((((...(...(((((((((((.(((.........))).))))))..)))))...)..)).............(((((((.......))))))).......))..))))..... ( -38.40) >DroSec_CAF1 38434 116 - 1 AGCGAGCCGCAAGCAAAUGUCCGCAGUCAGCCCAUA---UUGGCCGACUGCCC-GACAAACGAGGCAGAUAAAAAACGAUUGCAGCGAUACGAGCUGUAAAUAAAGAGGACCCUGGGAUC .(((..((....).....)..)))......((((..---..((((..((((((-(.....)).)))))...........(((((((.......))))))).......)).)).))))... ( -33.60) >DroSim_CAF1 17512 117 - 1 AGCGAGCCGCAAGCAAAUGUCCGCAGUCAGCCCAUA---UUGGCCGACUGCCCGGACAAACGAGGCAGAUAAAAAACGAUUGCAGCGAUACGAGCUGUAAAUAAAGAGGACCCUGGGAUC .....((((....)...(((((((((((.(((....---..))).))))))..))))).....))).............(((((((.......))))))).................... ( -35.30) >DroEre_CAF1 19923 117 - 1 ACCAAGCCGCAAGCGAGUGUCCGCAGUCAGCCCAUA---UUGGCCGACUGCCCGGACAAACGAGGCAGAUAAAAAACGAUUGCAGCGAUACGAGCUGUAAAUAAAGCGGACCCUGGGAUC .(((..((((...((..(((((((((((.(((....---..))).))))))..)))))..)).................(((((((.......))))))).....))))....))).... ( -43.80) >DroYak_CAF1 5952 117 - 1 AGCAAGCGGCAAGCGAAUGUCCGCAGUCAGUUCAUA---UUGGCCGACUGUCCGGACAAACGAGGCAGAUAAAAAACUAUUGCAGCGAUACGAGCUGUAAAUAAAGGGAAACCUGGUAUC ........((...((..(((((((((((.(((....---..))).)))))..))))))..))..)).((((........(((((((.......)))))))....(((....)))..)))) ( -36.40) >consensus AGCAAGCCGCAAGCAAAUGUCCGCAGUCAGCCCAUA___UUGGCCGACUGCCCGGACAAACGAGGCAGAUAAAAAACGAUUGCAGCGAUACGAGCUGUAAAUAAAGAGGACCCUGGGAUC .....((((....)...(((((((((((.(((.........))).))))))..))))).....))).............(((((((.......))))))).................... (-31.18 = -31.10 + -0.08)

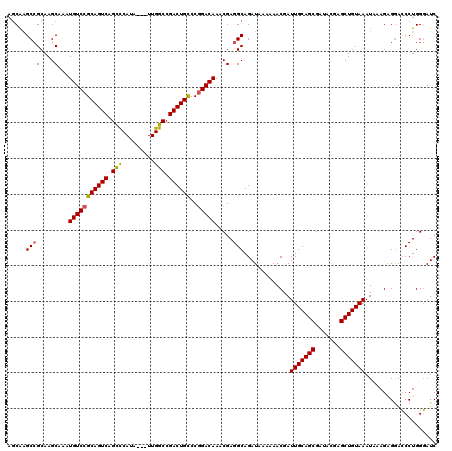
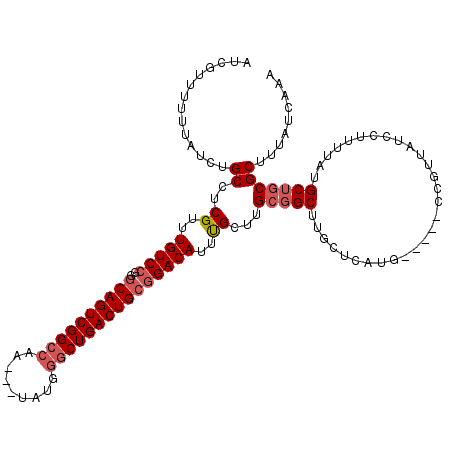
| Location | 1,319,936 – 1,320,051 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.97 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -27.73 |
| Energy contribution | -28.69 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1319936 115 + 20766785 AUCGUUUUUUAUCUGCCUCGUUUGUCCGGGCAGUCGGCCAAUAAUAUGGGCUGACUGCGGACAUUUGCUUGCGGCCUGCUCAUG-----CCGUUAUCCUUUUAUGCUCCGCUUUAUCAAA ..............(((.((......)))))((((((((.........))))))))((((((((......(((((........)-----)))).........))).)))))......... ( -34.96) >DroSec_CAF1 38474 111 + 1 AUCGUUUUUUAUCUGCCUCGUUUGUC-GGGCAGUCGGCCAA---UAUGGGCUGACUGCGGACAUUUGCUUGCGGCUCGCUCAUG-----CCGUUAUCCUUUUAUGCUGCGCUUUAUCAAA ..............(((.((.....)-))))((((((((..---....))))))))((((.(((......(((((........)-----)))).........)))))))........... ( -34.56) >DroSim_CAF1 17552 112 + 1 AUCGUUUUUUAUCUGCCUCGUUUGUCCGGGCAGUCGGCCAA---UAUGGGCUGACUGCGGACAUUUGCUUGCGGCUCGCUCAUG-----CCGUUAUCCUUUUAUGCUGCGCUUUAUCAAA ..............((..((..(((((..((((((((((..---....)))))))))))))))..))...(((((........)-----))))..............))........... ( -36.00) >DroEre_CAF1 19963 116 + 1 AUCGUUUUUUAUCUGCCUCGUUUGUCCGGGCAGUCGGCCAA---UAUGGGCUGACUGCGGACACUCGCUUGCGGCUUGGUCAUCCUUAUCCGUUAUCCUU-UGUGCUGCGCUUUAUCAAA ..............((..((..(((((..((((((((((..---....)))))))))))))))..))...(((((..((.................))..-...)))))))......... ( -36.23) >DroYak_CAF1 5992 112 + 1 AUAGUUUUUUAUCUGCCUCGUUUGUCCGGACAGUCGGCCAA---UAUGAACUGACUGCGGACAUUCGCUUGCCGCUUGCUUAUG-----CCGUUAUCCUUUUAUGCUGCGCCUUAUCAAA ..............((..((..((((((..(((((((....---......)))))))))))))..))...))(((..((.....-----...............)).))).......... ( -24.25) >consensus AUCGUUUUUUAUCUGCCUCGUUUGUCCGGGCAGUCGGCCAA___UAUGGGCUGACUGCGGACAUUUGCUUGCGGCUUGCUCAUG_____CCGUUAUCCUUUUAUGCUGCGCUUUAUCAAA ..............((..((..(((((..((((((((((.........)))))))))))))))..))...(((((.............................)))))))......... (-27.73 = -28.69 + 0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:25:18 2006