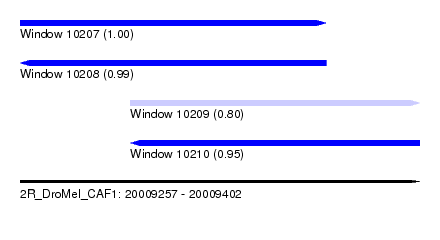
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,009,257 – 20,009,402 |
| Length | 145 |
| Max. P | 0.996469 |

| Location | 20,009,257 – 20,009,368 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.75 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -25.26 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.996469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20009257 111 + 20766785 UAGGGGGAUGCACUGAGAGAACAGUUCUUAACUUUUUCGAAAGGGCAGGAAAAAGUAUGCAUGAGUAUUUUGUGGUUUGCAACUCAGCUCAUUCACUGCAUACUUUUCCCU-------- ...((((((((.((..(((((.(((.....))))))))...)).)))....(((((((((((((((...((((.....))))....))))).....)))))))))))))))-------- ( -31.60) >DroSec_CAF1 26019 111 + 1 UAAGGGGCUUCACUGAGAGAACAGUUGCUAACUUUUUCGAAAGGGAAGCAAAAAGUAUGCAAGAGUAUUUUGUGGUUCGCAACUCUGCUCUCUCAUUGCAUACUUUUCCCU-------- ..(((((((((.((..(((((.(((.....))))))))...)).))))).(((((((((((((((((..(((((...)))))...)))))).....)))))))))))))))-------- ( -37.50) >DroSim_CAF1 26191 111 + 1 CAAGGGGCUGCACUGAGAGAACAGUUCCUAACUUUUUCGAAUGAGCAGCAAAAAGUAUGCAAGAGUAUUUUGUGGUUCGCAACUCUGCUCUCUCAUUGCAUACUUUUUCCU-------- ...((((((((.(...(((((.(((.....))))))))....).)))))((((((((((((((((((..(((((...)))))...)))))).....)))))))))))))))-------- ( -35.10) >DroEre_CAF1 31961 110 + 1 UG-GGGGCUGCACUGAGGGAAUAGUUCCCAACUUCUUUGCUGGCACAGCAAAAAGUAUGCAAGGGUAUUUUGUGGUUCGCUACCCAGCUCCUUCACUGCAUACUUUUCCCC-------- .(-(((((((((..((((((.....))))....))..))).)))......(((((((((((((((....(((.(((.....))))))..))))...)))))))))))))))-------- ( -36.70) >DroYak_CAF1 31349 118 + 1 UA-GUGGCUGCACUGAGGAAAUAGUUCUGCACUUCUUUGAAAGCACAGCAAAAAGUAUGCAAGAGUGUUUUGUGGUGUGCCACCCAAGUUAUUCACUGCAUACUUUUUCCCUUUUCUCA .(-(((....))))(((((((..(((.(((............))).)))((((((((((((.(((((.((((.((((...)))))))).)))))..))))))))))))...))))))). ( -36.10) >consensus UA_GGGGCUGCACUGAGAGAACAGUUCCUAACUUUUUCGAAAGCGCAGCAAAAAGUAUGCAAGAGUAUUUUGUGGUUCGCAACUCAGCUCAUUCACUGCAUACUUUUCCCU________ ...((((((((.((..(((((.(((.....))))))))...)).))))).((((((((((((((((...((((.....))))....))))).....))))))))))))))......... (-25.26 = -25.90 + 0.64)

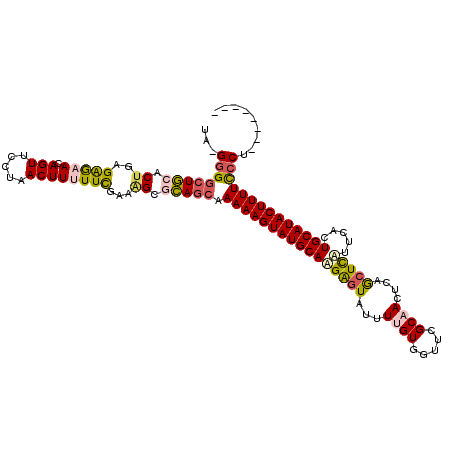
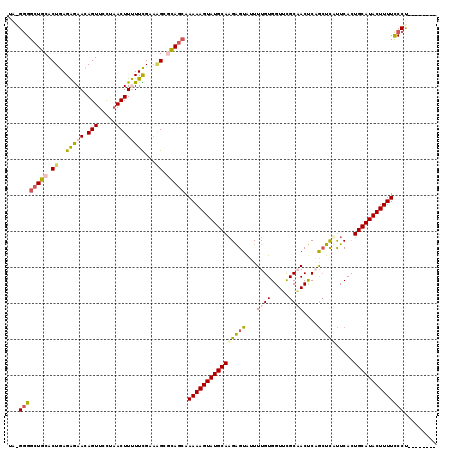
| Location | 20,009,257 – 20,009,368 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.75 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -20.75 |
| Energy contribution | -20.19 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20009257 111 - 20766785 --------AGGGAAAAGUAUGCAGUGAAUGAGCUGAGUUGCAAACCACAAAAUACUCAUGCAUACUUUUUCCUGCCCUUUCGAAAAAGUUAAGAACUGUUCUCUCAGUGCAUCCCCCUA --------(((((((((((((((((......))(((((...............))))))))))))))))))))((.((((....))))......((((......))))))......... ( -27.26) >DroSec_CAF1 26019 111 - 1 --------AGGGAAAAGUAUGCAAUGAGAGAGCAGAGUUGCGAACCACAAAAUACUCUUGCAUACUUUUUGCUUCCCUUUCGAAAAAGUUAGCAACUGUUCUCUCAGUGAAGCCCCUUA --------((((....((...((.(((((((((((((((((((..............))))).)))..(((((...((((....))))..)))))))))))))))).))..)))))).. ( -31.64) >DroSim_CAF1 26191 111 - 1 --------AGGAAAAAGUAUGCAAUGAGAGAGCAGAGUUGCGAACCACAAAAUACUCUUGCAUACUUUUUGCUGCUCAUUCGAAAAAGUUAGGAACUGUUCUCUCAGUGCAGCCCCUUG --------((((((((((((((((.(((...((......))(.....)......))))))))))))))))(((((...(((..........)))((((......))))))))).))).. ( -30.70) >DroEre_CAF1 31961 110 - 1 --------GGGGAAAAGUAUGCAGUGAAGGAGCUGGGUAGCGAACCACAAAAUACCCUUGCAUACUUUUUGCUGUGCCAGCAAAGAAGUUGGGAACUAUUCCCUCAGUGCAGCCCC-CA --------(((((((((((((((((......)))(((((.............)))))..)))))))))).((((..(.(((......)))((((.....))))...)..)))))))-). ( -40.82) >DroYak_CAF1 31349 118 - 1 UGAGAAAAGGGAAAAAGUAUGCAGUGAAUAACUUGGGUGGCACACCACAAAACACUCUUGCAUACUUUUUGCUGUGCUUUCAAAGAAGUGCAGAACUAUUUCCUCAGUGCAGCCAC-UA .(((.(((.(((((((((((((((.((.....(((((((...)))).))).....))))))))))))))).(((..((((....))))..)))..)).))).)))((((....)))-). ( -35.90) >consensus ________AGGGAAAAGUAUGCAGUGAAAGAGCUGAGUUGCGAACCACAAAAUACUCUUGCAUACUUUUUGCUGCCCUUUCGAAAAAGUUAGGAACUGUUCUCUCAGUGCAGCCCC_UA .........(((((((((((((((..........((((...............)))))))))))))))))(((((...........(((.....)))...........))))))).... (-20.75 = -20.19 + -0.56)

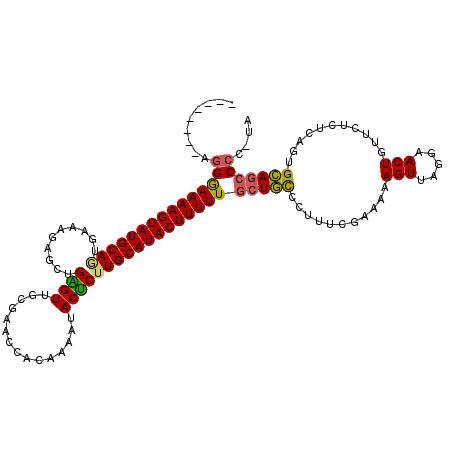
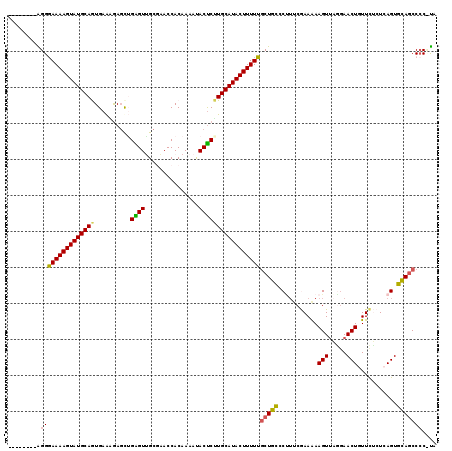
| Location | 20,009,297 – 20,009,402 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -31.06 |
| Consensus MFE | -17.24 |
| Energy contribution | -17.96 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20009297 105 + 20766785 AAGGGCAGGAAAAAGUAUGCAUGAGUAUUUUGUGGUUUGCAACUCAGCUCAUUCACUGCAUACUUUUCCCU--------GUGUAGGCAGUGCUCUUCCUCACCGGGUUAUGUA ..(((((((.((((((((((((((((...((((.....))))....))))).....))))))))))).)))--------))(.(((.((....)).)))).)).......... ( -32.10) >DroSec_CAF1 26059 105 + 1 AAGGGAAGCAAAAAGUAUGCAAGAGUAUUUUGUGGUUCGCAACUCUGCUCUCUCAUUGCAUACUUUUCCCU--------GUGUAGACAGUGCUCUUCCUCACCGGGUUAUGUA .((((((((((((((((((((((((((..(((((...)))))...)))))).....)))))))))))..((--------((....)))))))).))))).............. ( -30.60) >DroSim_CAF1 26231 105 + 1 AUGAGCAGCAAAAAGUAUGCAAGAGUAUUUUGUGGUUCGCAACUCUGCUCUCUCAUUGCAUACUUUUUCCU--------GUGUAGGCAGUGCUCUUCCUCACCUGGUUAUGUA ((((.(((.((((((((((((((((((..(((((...)))))...)))))).....))))))))))))...--------(((.(((.((....)).))))))))).))))... ( -31.30) >DroEre_CAF1 32000 105 + 1 UGGCACAGCAAAAAGUAUGCAAGGGUAUUUUGUGGUUCGCUACCCAGCUCCUUCACUGCAUACUUUUCCCC--------GUGCAGCGAGUGCUUCUGCUCACCGCGUUAUGUA ..((((.(..(((((((((((((((....(((.(((.....))))))..))))...)))))))))))..).--------)))).(((.(((........))))))........ ( -32.70) >DroYak_CAF1 31388 113 + 1 AAGCACAGCAAAAAGUAUGCAAGAGUGUUUUGUGGUGUGCCACCCAAGUUAUUCACUGCAUACUUUUUCCCUUUUCUCAGUGUACUUAGUACUUUUCCUCACCGCGUUAUGUA ..(((.(((((((((((((((.(((((.((((.((((...)))))))).)))))..)))))))))))).............((((...)))).............))).))). ( -28.60) >consensus AAGCGCAGCAAAAAGUAUGCAAGAGUAUUUUGUGGUUCGCAACUCAGCUCAUUCACUGCAUACUUUUCCCU________GUGUAGGCAGUGCUCUUCCUCACCGGGUUAUGUA ....(((...((((((((((((((((...((((.....))))....))))).....)))))))))))............(((.(((.((....)).)))))).......))). (-17.24 = -17.96 + 0.72)

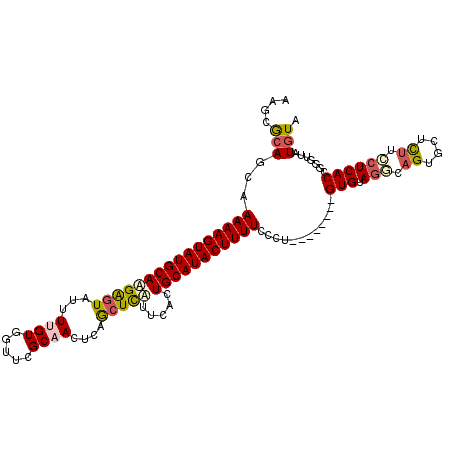
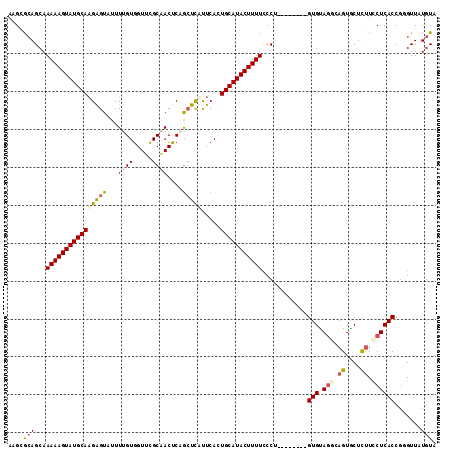
| Location | 20,009,297 – 20,009,402 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -20.04 |
| Energy contribution | -19.04 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20009297 105 - 20766785 UACAUAACCCGGUGAGGAAGAGCACUGCCUACAC--------AGGGAAAAGUAUGCAGUGAAUGAGCUGAGUUGCAAACCACAAAAUACUCAUGCAUACUUUUUCCUGCCCUU .........(((((........)))))......(--------(((((((((((((((((......))(((((...............)))))))))))))))))))))..... ( -30.56) >DroSec_CAF1 26059 105 - 1 UACAUAACCCGGUGAGGAAGAGCACUGUCUACAC--------AGGGAAAAGUAUGCAAUGAGAGAGCAGAGUUGCGAACCACAAAAUACUCUUGCAUACUUUUUGCUUCCCUU .............((((..((((.((((....))--------)).(((((((((((((.(((...((......))(.....)......)))))))))))))))))))).)))) ( -28.70) >DroSim_CAF1 26231 105 - 1 UACAUAACCAGGUGAGGAAGAGCACUGCCUACAC--------AGGAAAAAGUAUGCAAUGAGAGAGCAGAGUUGCGAACCACAAAAUACUCUUGCAUACUUUUUGCUGCUCAU ...........((((((.((.((...)))).).(--------((.(((((((((((((.(((...((......))(.....)......)))))))))))))))).)))))))) ( -28.60) >DroEre_CAF1 32000 105 - 1 UACAUAACGCGGUGAGCAGAAGCACUCGCUGCAC--------GGGGAAAAGUAUGCAGUGAAGGAGCUGGGUAGCGAACCACAAAAUACCCUUGCAUACUUUUUGCUGUGCCA ........((((((((........))))))))((--------((.((((((((((((((......)))(((((.............)))))..))))))))))).)))).... ( -39.42) >DroYak_CAF1 31388 113 - 1 UACAUAACGCGGUGAGGAAAAGUACUAAGUACACUGAGAAAAGGGAAAAAGUAUGCAGUGAAUAACUUGGGUGGCACACCACAAAACACUCUUGCAUACUUUUUGCUGUGCUU .......((((((........((((...)))).............(((((((((((((.((.....(((((((...)))).))).....)))))))))))))))))))))... ( -28.80) >consensus UACAUAACCCGGUGAGGAAGAGCACUGCCUACAC________AGGGAAAAGUAUGCAGUGAAAGAGCUGAGUUGCGAACCACAAAAUACUCUUGCAUACUUUUUGCUGCCCUU ..........((((........))))................((.(((((((((((((..........((((...............))))))))))))))))).))...... (-20.04 = -19.04 + -1.00)

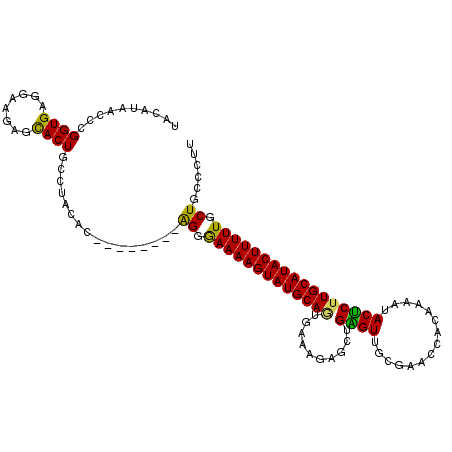
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:00 2006