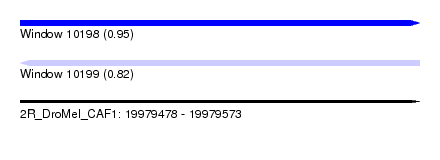
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,979,478 – 19,979,573 |
| Length | 95 |
| Max. P | 0.951362 |

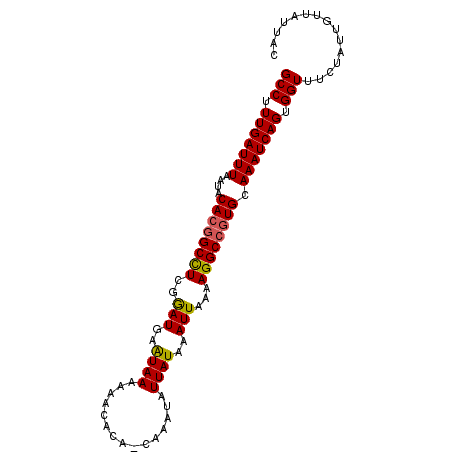
| Location | 19,979,478 – 19,979,573 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 92.70 |
| Mean single sequence MFE | -19.86 |
| Consensus MFE | -19.50 |
| Energy contribution | -19.44 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
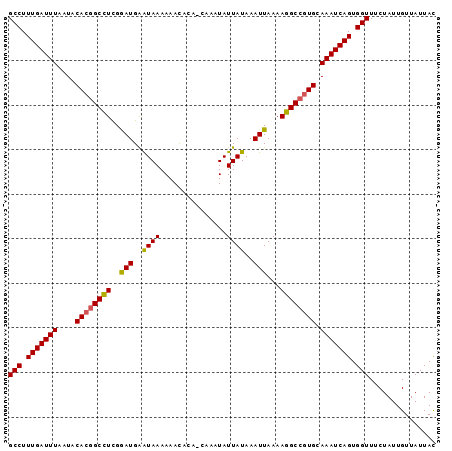
>2R_DroMel_CAF1 19979478 95 + 20766785 GCCUUUGAUUUAAUACACGGCCUCGGAUGAGUAAAAAAUACA-CAAAUAUUAUAAAUUAAAAGGCCGUGCAAAUCAGUGGUUUCUAUUGUUAUUAC (((.(((((((....((((((((..(((..((((........-......))))..)))...)))))))).))))))).)))............... ( -22.24) >DroSec_CAF1 20490 96 + 1 GCCUUUGAUUUAAUACACGGCCUCGGAUGAAUAAAAAACACAGCAAAUAUUAUAAAUUAAAAGGCCGUGCAAAUCAGUGGUUUCUAUUGUUAUUAU (((.(((((((....((((((((..(((..((((...............))))..)))...)))))))).))))))).)))............... ( -21.86) >DroSim_CAF1 21176 96 + 1 GCCUUUGAUUUAAUUCACGGCCUCGGAUGAAUAAAAAACACAGCAAAUAUUAUAAAUUAAAAGGCCGUGCAAAUCAGUGGUUUCUAUUGUUAUUAC (((.(((((((....((((((((..(((..((((...............))))..)))...)))))))).))))))).)))............... ( -21.86) >DroEre_CAF1 20574 91 + 1 GCCUUUGAUUUAAUGCA--GCUUUGAAUGGAUAAAAAACA---CAAAUAUUAUAAAUUAAAAGGCCGUGCAAAUCAGUGGUUUCUAUUGUUAUUAC (((.((((((...((((--(((((.(((..((((......---......))))..)))..)))))..)))))))))).)))............... ( -13.50) >consensus GCCUUUGAUUUAAUACACGGCCUCGGAUGAAUAAAAAACACA_CAAAUAUUAUAAAUUAAAAGGCCGUGCAAAUCAGUGGUUUCUAUUGUUAUUAC (((.(((((((....((((((((..(((..((((...............))))..)))...)))))))).))))))).)))............... (-19.50 = -19.44 + -0.06)

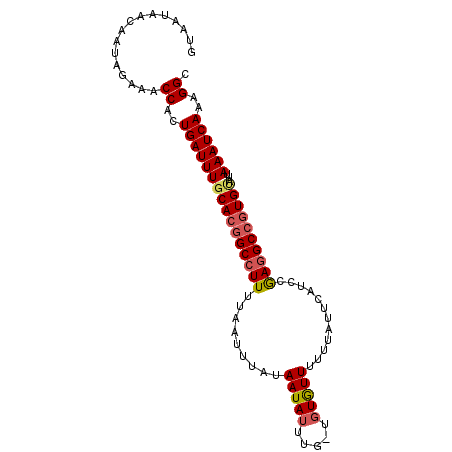
| Location | 19,979,478 – 19,979,573 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 92.70 |
| Mean single sequence MFE | -19.33 |
| Consensus MFE | -17.50 |
| Energy contribution | -18.25 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19979478 95 - 20766785 GUAAUAACAAUAGAAACCACUGAUUUGCACGGCCUUUUAAUUUAUAAUAUUUG-UGUAUUUUUUACUCAUCCGAGGCCGUGUAUUAAAUCAAAGGC ................((..((((((((((((((((...............((-.(((.....))).))...)))))))))))...)))))..)). ( -22.77) >DroSec_CAF1 20490 96 - 1 AUAAUAACAAUAGAAACCACUGAUUUGCACGGCCUUUUAAUUUAUAAUAUUUGCUGUGUUUUUUAUUCAUCCGAGGCCGUGUAUUAAAUCAAAGGC ................((..((((((((((((((((......((.(((((.....)))))...)).......)))))))))))...)))))..)). ( -22.72) >DroSim_CAF1 21176 96 - 1 GUAAUAACAAUAGAAACCACUGAUUUGCACGGCCUUUUAAUUUAUAAUAUUUGCUGUGUUUUUUAUUCAUCCGAGGCCGUGAAUUAAAUCAAAGGC ................((..((((((((((((((((......((.(((((.....)))))...)).......)))))))))...)))))))..)). ( -21.62) >DroEre_CAF1 20574 91 - 1 GUAAUAACAAUAGAAACCACUGAUUUGCACGGCCUUUUAAUUUAUAAUAUUUG---UGUUUUUUAUCCAUUCAAAGC--UGCAUUAAAUCAAAGGC ................((..(((((((..((((.(((.(((..((((......---......))))..))).)))))--))...)))))))..)). ( -10.20) >consensus GUAAUAACAAUAGAAACCACUGAUUUGCACGGCCUUUUAAUUUAUAAUAUUUG_UGUGUUUUUUAUUCAUCCGAGGCCGUGUAUUAAAUCAAAGGC ................((..((((((((((((((((.........(((((.....)))))............))))))))))...))))))..)). (-17.50 = -18.25 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:49 2006