
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,979,152 – 19,979,256 |
| Length | 104 |
| Max. P | 0.572718 |

| Location | 19,979,152 – 19,979,256 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.18 |
| Mean single sequence MFE | -38.03 |
| Consensus MFE | -11.32 |
| Energy contribution | -13.07 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19979152 104 + 20766785 CUUAGCCCGGUCG---GG-AUGUCCUUCAGCUGACGUCCUC-----GUUGGCGGCUGAGUGG----GGGCCAUUAAUGCAUCGGAUUAACCCAACCAUAUGCAGAUAUG---AGUAUAUC ........(((.(---((-..((((....((..(((....)-----))..)).((..(((((----...)))))...))...))))...))).)))((((((.......---.)))))). ( -32.40) >DroSec_CAF1 20166 104 + 1 CUUGGCCCGGUCG---GG-UUGUCCUUCAGCUGACGUCCUC-----GUAGGCGGCUGAGUGG----CGGCCAUUAAAGCAUCGGACUAACCCGACCAUAUGCAUCUACG---GGUGUAUC ........(((((---((-((((((....(((.(((....)-----)).)))(((((.....----)))))...........)))).)))))))))..(((((((....---))))))). ( -47.30) >DroSim_CAF1 20852 104 + 1 CUUGGCCCGGUCG---GG-AUGUCCUUCAGCUGACGUCCUC-----GUAGGCGGCUGAGUGG----CGGCCAUUAAUGCAUCGGAUUAACCCGACCAUAUGCAUCUACG---GGUGUAUC ........(((((---((-..((((....(((.(((....)-----)).)))(((((.....----)))))...........))))...)))))))..(((((((....---))))))). ( -42.80) >DroEre_CAF1 20260 103 + 1 CUUGGCCCGCUCG---GG-AUGUCCUUCAGCUGACAUCCUUCAGCUGACGUCGCCUGAGUGG----AGGCCAUCAAUGCAGCGGAUCAGACCGACUAUAUGCAUAUAG---------AUG ..(((((((((((---((-.((.(..((((((((......)))))))).).)))))))))).----.)))))...(((((.(((......)))......)))))....---------... ( -41.40) >DroYak_CAF1 20819 116 + 1 CCUGGCCCGCUCG---GGAAUGUCCUUCAGCCGACAUCCUUCAGCUGACGUGG-CUGAGUGGAGGGAGGCCAUUAAUGCAUCGAAUCAGCCCGACUAUAUGCAUAUAUAUCUACUAUAUC ..((((((.((((---(((.((((........))))))))((((((.....))-))))...))).).)))))...((((((....((.....))....))))))................ ( -34.70) >DroAna_CAF1 19183 95 + 1 -----CGAGGUGAAGCUG-AUGCCCUUCAGUUGAUGUCCUC-----AUAAUCGGCUGAGUGG----AGGACAUUAAUGCAUCAGAUUAUGU-------UUGUAAGUCCC---UACAUAUA -----..(((.((..(((-((((......((((((((((((-----....((....))...)----)))))))))))))))))).((((..-------..)))).))))---)....... ( -29.60) >consensus CUUGGCCCGGUCG___GG_AUGUCCUUCAGCUGACGUCCUC_____GUAGGCGGCUGAGUGG____AGGCCAUUAAUGCAUCGGAUUAACCCGACCAUAUGCAUAUACG___AGUAUAUC ..(((((................((((((((((.(..............).)))))))).)).....)))))....((((((((......)))).....))))................. (-11.32 = -13.07 + 1.75)

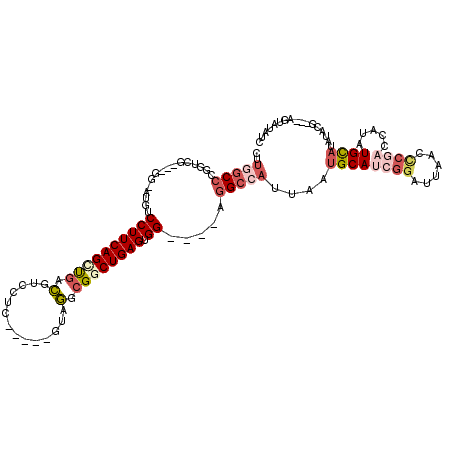
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:47 2006