| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,975,035 – 19,975,143 |
| Length | 108 |
| Max. P | 0.594197 |

| Location | 19,975,035 – 19,975,143 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.99 |
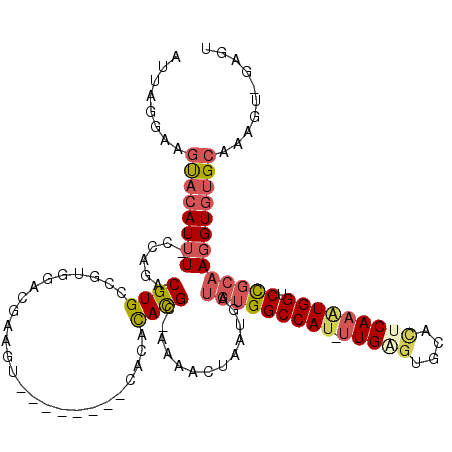
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -16.61 |
| Energy contribution | -17.64 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
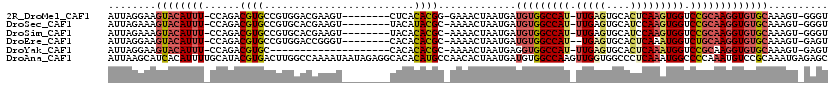
>2R_DroMel_CAF1 19975035 108 - 20766785 AUUAGGAAGUACAUUU-CCAGACGUGCCGUGGACGAAGU--------CUCACACGG-GAAACUAAUGAUGUGGCCAU-UUGAGUGCACUCAAGUGGUCCGCAAGGUGUGCAAAGU-GGGU ....(((((....)))-)).(.((..((((((((...))--------).)))..((-....)).....(((((((((-(((((....))))))))).))))).))..))).....-.... ( -38.20) >DroSec_CAF1 16097 108 - 1 AUUAGAAAGUACAUUU-CCAGACGUGCCGUGCACGAAGU--------UACAUACGC-AAAACUAAUGAUGUGGCCAU-UUGAGUGCAUCCAAGUGGUCCGCAAGGUGUGCAAAGU-GGGU ........((((((((-.....(((((...)))))....--------.........-...........(((((((((-(((........))))))).))))))))))))).....-.... ( -29.30) >DroSim_CAF1 16730 108 - 1 AUUAGAAAGUACAUUU-CCAGACGUGCCGUGCACGAAGU--------UACACACGC-AAAACUAAUGAUGUGGCCAU-UUGAGUGCAUCCAAGUGGUCCGCAAGGUGUGCAAAGU-GGGU ...............(-(((..(((((...)))))....--------..(((((..-....(....).(((((((((-(((........))))))).)))))..))))).....)-))). ( -29.50) >DroEre_CAF1 16159 107 - 1 AUUAGGAAGUACAUUU-CCAGACGUGCCGUGGACCGGGU--------CACACACGC-AAAACUAAUGAUGUGGCCAU--UGAGUGCACUCAAAUGGUCUGCAAGGUGUGCAAAGU-GAGU ....(((((....)))-)).(.((..(((..((((((((--------((((((...-........)).))))))).(--((((....))))).)))))..)..))..))).....-.... ( -34.20) >DroYak_CAF1 16760 96 - 1 AUUAGGAAGUACAUUU-CCAGACGUGC--------------------CACACACGC-AAAACUAAUGAGGUGGCCAU-UUGAGUGCACUCAAAUGGUCCGCAAGGUGUGCAAAGU-GAGU ........((((((((-...(.((((.--------------------....)))))-............((((((((-(((((....))))))))).)))).)))))))).....-.... ( -30.90) >DroAna_CAF1 15462 120 - 1 AUUAAGCAUCACAUUUUGCAUACGUGACUUGGCCAAAAUAAUAGAGGCACACAUGCCAACACUAAUGAUGUGGCCAAGUUGGUGGCCCUCAAAUGGCCCCAAAUGUCCGCAAAUGAGAGC .....((.((.(((((.((..((((((((((((((...((.(((.((((....))))....))).))...))))))))))((.((((.......))))))..))))..))))))).)))) ( -40.30) >consensus AUUAGGAAGUACAUUU_CCAGACGUGCCGUGGACGAAGU________CACACACGC_AAAACUAAUGAUGUGGCCAU_UUGAGUGCACUCAAAUGGUCCGCAAGGUGUGCAAAGU_GAGU ........((((((((......((((.........................)))).............(((((((((.(((((....))))))))).))))))))))))).......... (-16.61 = -17.64 + 1.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:41 2006