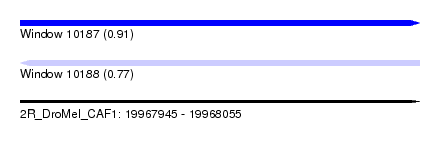
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,967,945 – 19,968,055 |
| Length | 110 |
| Max. P | 0.908836 |

| Location | 19,967,945 – 19,968,055 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.04 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -17.66 |
| Energy contribution | -17.71 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
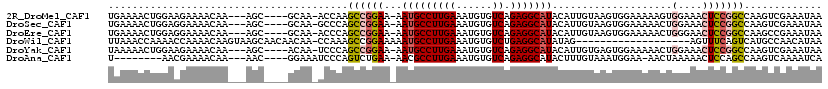
>2R_DroMel_CAF1 19967945 110 + 20766785 UUAUUUCGACUUGGCCGGAGUUUCCACUUUUUCCACUUACAAUGUAUGCCUCUGACACAUUUCAAGGCAUU-UUCCGGCUUGGU-UUGC----GCU---UUGUUUUCUUCCAGUUUUCA .......((((.((((((((..............((.......))((((((.(((......))))))))).-)))))))).)))-)...----...---.................... ( -24.00) >DroSec_CAF1 9088 110 + 1 UUAUUUCGACUUGGCCGGAGUUUCCAGUUUUUCCACUUACAAUGUAUGCCUCUGACACAUUUCAAGGCAUU-UUCCGGCUGGGC-UUGC----GCU---UUGUUUUCCUCCAGUUUUCA ......(((((..(((((((.....(((......)))........((((((.(((......))))))))).-)))))))..)).-))).----...---.................... ( -25.40) >DroEre_CAF1 8974 110 + 1 UUAUUUCGGCUUGGCCGGAGUUCCCAGUUUUUCCACUUACAAUGUAUGCCUCUGACACAUUUCAAGGCAUU-UUCCGGCUGGGU-UUGC----GCU---UUGUUUUCCUCCAGUUUUCA .......((((..(((((((.....(((......)))........((((((.(((......))))))))).-)))))))..)))-)...----...---.................... ( -26.70) >DroWil_CAF1 14048 99 + 1 UUAUGUUGGCAUGACUGAAACU-------------------CUAUAUGCCUCAGACACAUUUCAAGGCAUUUUUCCGGCUUUGG-UUGUUGUUGCUUACUUGUUUUGGUUUUGGUUUAA .........((.(((..((((.-------------------......((..(((.((....(((((((.........)))))))-.)))))..))......))))..))).))...... ( -19.32) >DroYak_CAF1 9138 110 + 1 UUAUUUCGACUUGGCCGGAGUUUCCAGUUUUUCCACUCACAAUGUAUGCCUCUGACACAUUUCAAGGCAUU-UUCCGGCUGGGA-UUGU----GCU---UUGUUUUCUUCCAGUUUUUA ......(((((..(((((((.....(((......)))........((((((.(((......))))))))).-)))))))..)).-))).----...---.................... ( -25.60) >DroAna_CAF1 8823 102 + 1 UGAUUUUGACUUGGCUGGAGUUUUUAGUU-UUCCAUUUACAAAGUAUGCCUCUGACACAUUUCAAGGCGUU-UUCAGACUGGGAUUUCC----GUU---UUGUUUUCGUU--------A .((....(((..(((.((((.((((((((-(..............((((((.(((......))))))))).-...))))))))).))))----)))---..))).))...--------. ( -20.87) >consensus UUAUUUCGACUUGGCCGGAGUUUCCAGUUUUUCCACUUACAAUGUAUGCCUCUGACACAUUUCAAGGCAUU_UUCCGGCUGGGA_UUGC____GCU___UUGUUUUCCUCCAGUUUUCA .........((..(((((((.........................((((((.(((......)))))))))..)))))))..)).................................... (-17.66 = -17.71 + 0.06)

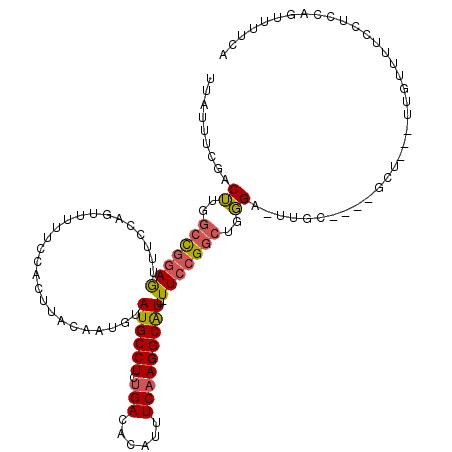
| Location | 19,967,945 – 19,968,055 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.04 |
| Mean single sequence MFE | -21.23 |
| Consensus MFE | -14.16 |
| Energy contribution | -14.75 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19967945 110 - 20766785 UGAAAACUGGAAGAAAACAA---AGC----GCAA-ACCAAGCCGGAA-AAUGCCUUGAAAUGUGUCAGAGGCAUACAUUGUAAGUGGAAAAAGUGGAAACUCCGGCCAAGUCGAAAUAA ....................---...----....-.....((((((.-.(((((((((......))).))))))....................(....)))))))............. ( -21.60) >DroSec_CAF1 9088 110 - 1 UGAAAACUGGAGGAAAACAA---AGC----GCAA-GCCCAGCCGGAA-AAUGCCUUGAAAUGUGUCAGAGGCAUACAUUGUAAGUGGAAAAACUGGAAACUCCGGCCAAGUCGAAAUAA .((...((((((........---.((----....-))((((......-.(((((((((......))).)))))).(((.....)))......))))...)))))).....))....... ( -28.20) >DroEre_CAF1 8974 110 - 1 UGAAAACUGGAGGAAAACAA---AGC----GCAA-ACCCAGCCGGAA-AAUGCCUUGAAAUGUGUCAGAGGCAUACAUUGUAAGUGGAAAAACUGGGAACUCCGGCCAAGCCGAAAUAA ........(((((....)..---...----....-.(((((......-.(((((((((......))).)))))).(((.....)))......)))))..))))(((...)))....... ( -24.90) >DroWil_CAF1 14048 99 - 1 UUAAACCAAAACCAAAACAAGUAAGCAACAACAA-CCAAAGCCGGAAAAAUGCCUUGAAAUGUGUCUGAGGCAUAUAG-------------------AGUUUCAGUCAUGCCAACAUAA .................((((...(((.......-((......)).....)))))))....(((.(((((((......-------------------.))))))).))).......... ( -14.40) >DroYak_CAF1 9138 110 - 1 UAAAAACUGGAAGAAAACAA---AGC----ACAA-UCCCAGCCGGAA-AAUGCCUUGAAAUGUGUCAGAGGCAUACAUUGUGAGUGGAAAAACUGGAAACUCCGGCCAAGUCGAAAUAA ......(((((......((.---..(----((((-(((.....))).-.(((((((((......))).))))))...)))))..))........(....)))))).............. ( -23.70) >DroAna_CAF1 8823 102 - 1 U--------AACGAAAACAA---AAC----GGAAAUCCCAGUCUGAA-AACGCCUUGAAAUGUGUCAGAGGCAUACUUUGUAAAUGGAA-AACUAAAAACUCCAGCCAAGUCAAAAUCA .--------...((..((((---(.(----(((........))))..-...(((((((......))).))))....)))))...((((.-..........))))............)). ( -14.60) >consensus UGAAAACUGGAAGAAAACAA___AGC____GCAA_ACCCAGCCGGAA_AAUGCCUUGAAAUGUGUCAGAGGCAUACAUUGUAAGUGGAAAAACUGGAAACUCCGGCCAAGUCGAAAUAA ........................................((((((...(((((((((......)).)))))))....................(....)))))))............. (-14.16 = -14.75 + 0.59)
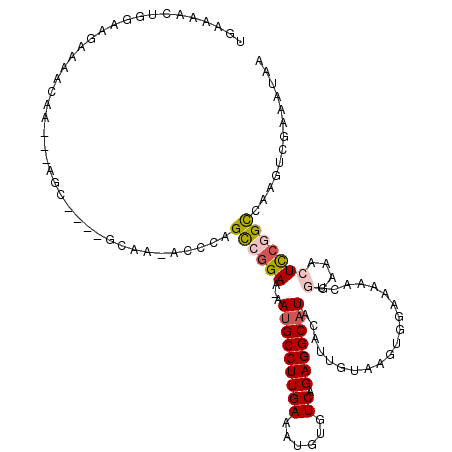
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:38 2006