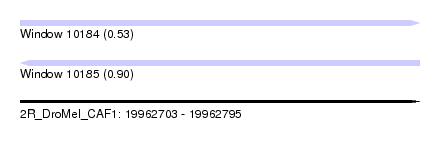
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,962,703 – 19,962,795 |
| Length | 92 |
| Max. P | 0.898935 |

| Location | 19,962,703 – 19,962,795 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -16.46 |
| Energy contribution | -17.21 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.49 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
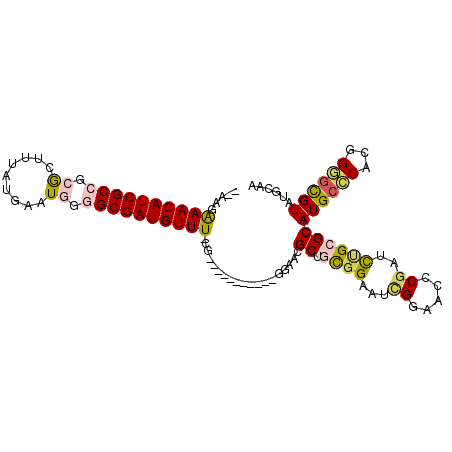
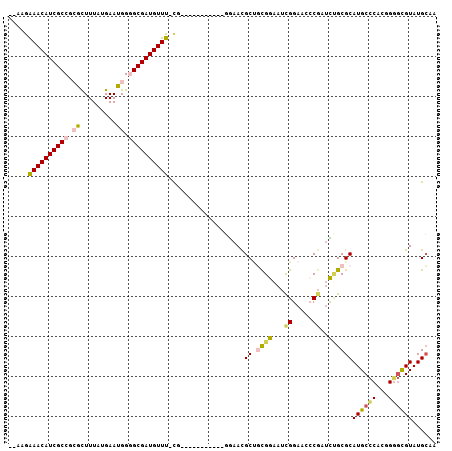
>2R_DroMel_CAF1 19962703 92 + 20766785 UUGCAUACGCACCGUGGGCAUGCGCAAAACGGGUUCCGAUUCCGCAGCAUUC------------CG-AAACAUCGCCCCAUUCAUAAAGCCCGGCGAUGUUUCUU-- .(((....)))((((..((....))...))))(((.((....)).)))....------------.(-((((((((((...............)))))))))))..-- ( -28.36) >DroVir_CAF1 31628 105 + 1 UUGCAUACAUGCGGAGCGCAUGCGCCAAACGUGUUCCAAUGCCACAGCGUUUCGCCUGGCCGUUAG-AAACAUCGCGCCAUUCAUAAAACCGGGCGAUGUUCUUGC- ..(((....)))(((((((...........)))))))...((((..(((...))).)))).((.((-((.((((((.((............)))))))))))).))- ( -32.10) >DroSec_CAF1 3861 93 + 1 UUGCAUACGCCCCGUGGGCAUGCGCAGAUCGGGUUCCGAUUCCGCAGCGUUCC-----------CG-AAACAUCGCCCCAUUCAUAAAGCGCGGCGAUGUUUCUU-- ..((....((((...)))).((((..((((((...)))))).)))))).....-----------.(-((((((((((.(...........).)))))))))))..-- ( -36.00) >DroSim_CAF1 3863 93 + 1 UUGCAUACGCCCCGUGGGCAUGCGCAGAUCGGGUUCCGAUUCCGCAGCGUUCC-----------CA-AAACAUCGCCCCAUUCAUAAAGCGCGGCGAUGUUUCUU-- ..((....((((...)))).((((..((((((...)))))).)))))).....-----------..-((((((((((.(...........).))))))))))...-- ( -32.00) >DroYak_CAF1 3828 95 + 1 UUGCAUACGCCCCGUGGGCAUGCGCGGAUCGGGUUCCGAUUCCAUGGCAUUCC-----------CG-AAACAUCGCCCCAUUCAUAAAGUGCGGCGAUGUUUCUUCC ..((((..((((...))))))))(((((((((...)))))))....)).....-----------.(-((((((((((.((((.....)))).))))))))))).... ( -38.20) >DroAna_CAF1 3665 94 + 1 UUUCAUACGCCCUGUGGGCAUGCGCGGUCGGUGUUCCGUUUCCGAUGCAUUCC-----------AGAAAACAUCGCACCAUUCAUAAAGUGCGGCGAUGUUUCUU-- ........((((...))))((((((((.(((....)))...))).)))))...-----------(((((.((((((..((((.....))))..))))))))))).-- ( -34.20) >consensus UUGCAUACGCCCCGUGGGCAUGCGCAGAUCGGGUUCCGAUUCCGCAGCAUUCC___________CG_AAACAUCGCCCCAUUCAUAAAGCGCGGCGAUGUUUCUU__ ..((((..((((...))))))))((.....((((....))))....))...................((((((((((...............))))))))))..... (-16.46 = -17.21 + 0.75)

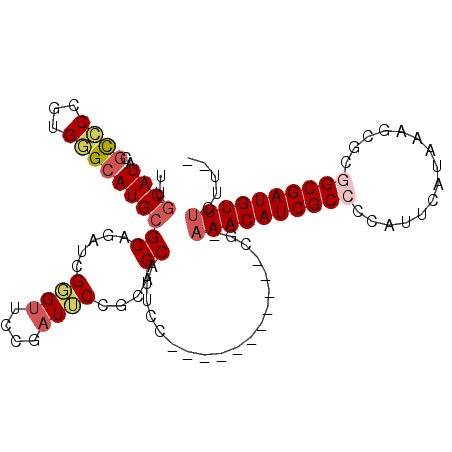
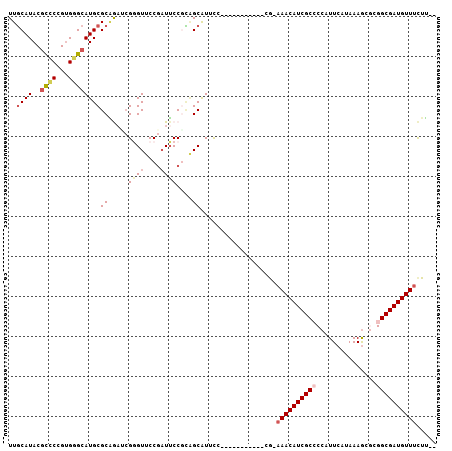
| Location | 19,962,703 – 19,962,795 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -37.29 |
| Consensus MFE | -22.52 |
| Energy contribution | -22.75 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.898935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19962703 92 - 20766785 --AAGAAACAUCGCCGGGCUUUAUGAAUGGGGCGAUGUUU-CG------------GAAUGCUGCGGAAUCGGAACCCGUUUUGCGCAUGCCCACGGUGCGUAUGCAA --..(((((((((((...............))))))))))-)(------------(.((((.((((((.(((...)))))))))))))..))....(((....))). ( -34.16) >DroVir_CAF1 31628 105 - 1 -GCAAGAACAUCGCCCGGUUUUAUGAAUGGCGCGAUGUUU-CUAACGGCCAGGCGAAACGCUGUGGCAUUGGAACACGUUUGGCGCAUGCGCUCCGCAUGUAUGCAA -(((.((((((((((((.((....)).))).)))))))))-......((((((((.....(((......)))....))))))))(((((((...))))))).))).. ( -38.10) >DroSec_CAF1 3861 93 - 1 --AAGAAACAUCGCCGCGCUUUAUGAAUGGGGCGAUGUUU-CG-----------GGAACGCUGCGGAAUCGGAACCCGAUCUGCGCAUGCCCACGGGGCGUAUGCAA --..(((((((((((.((.........)).))))))))))-)(-----------....)...(((((.((((...)))))))))((((((((....)).)))))).. ( -41.50) >DroSim_CAF1 3863 93 - 1 --AAGAAACAUCGCCGCGCUUUAUGAAUGGGGCGAUGUUU-UG-----------GGAACGCUGCGGAAUCGGAACCCGAUCUGCGCAUGCCCACGGGGCGUAUGCAA --..(((((((((((.((.........)).))))))))))-).-----------........(((((.((((...)))))))))((((((((....)).)))))).. ( -38.70) >DroYak_CAF1 3828 95 - 1 GGAAGAAACAUCGCCGCACUUUAUGAAUGGGGCGAUGUUU-CG-----------GGAAUGCCAUGGAAUCGGAACCCGAUCCGCGCAUGCCCACGGGGCGUAUGCAA (((.(((((((((((.((.........)).))))))))))-)(-----------((....((........))..)))..)))(((.((((((...)))))).))).. ( -42.00) >DroAna_CAF1 3665 94 - 1 --AAGAAACAUCGCCGCACUUUAUGAAUGGUGCGAUGUUUUCU-----------GGAAUGCAUCGGAAACGGAACACCGACCGCGCAUGCCCACAGGGCGUAUGAAA --......((((((((((((........))))).........(-----------((.((((..(((...(((....))).))).))))..)))...)))).)))... ( -29.30) >consensus __AAGAAACAUCGCCGCGCUUUAUGAAUGGGGCGAUGUUU_CG___________GGAACGCUGCGGAAUCGGAACCCGAUCUGCGCAUGCCCACGGGGCGUAUGCAA .....((((((((((.((.........)).))))))))))...................((.((((...((.....))..))))))((((((...))))))...... (-22.52 = -22.75 + 0.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:35 2006