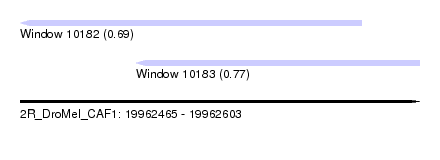
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,962,465 – 19,962,603 |
| Length | 138 |
| Max. P | 0.766895 |

| Location | 19,962,465 – 19,962,583 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 87.13 |
| Mean single sequence MFE | -30.97 |
| Consensus MFE | -26.43 |
| Energy contribution | -26.43 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
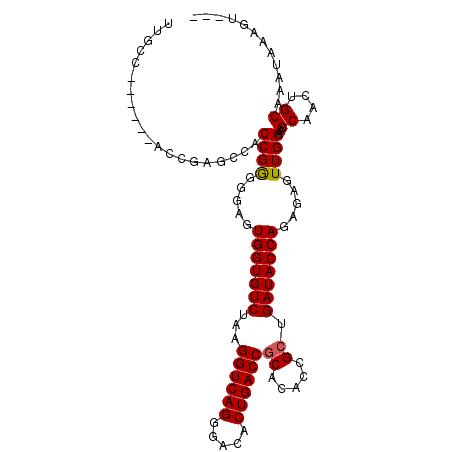
>2R_DroMel_CAF1 19962465 118 - 20766785 GAUUGGUGUCUAAGGUCAGGGACACUGACCGCACACCGCUGAUACCAGACAGUUGGAAGCAAGUGCGAAAAAAGUACGGAAACUAUAGUUUAUGAAAGUGACCAUCGUAUCUAGGAUC ..((((((((...((((((.....))))))((.....)).))))))))....(((((.((..((((.......))))(....)....(.((((....)))).)...)).))))).... ( -33.10) >DroSec_CAF1 3643 106 - 1 GAGUGGUGUCUAAGGUCAGGGACACUGACCGCAAACUGCUGAUACCAGAGAGUUGGAAGCAGGUGCAAAUAAAGU-----AACUAUAGUUUAUGAAAGUGACCAUCGUAUC------- ((.((((..((..((((((.....))))))(((..(((((....((((....)))).))))).)))..(((((.(-----(....)).)))))...))..)))))).....------- ( -29.50) >DroSim_CAF1 3637 106 - 1 GAGUGGUGUCUAAGGUCAGGGACACUGACCGCACACUGCUGAUACCAGAGAGUUGGAAGCAACUGCAAAUAAAGU-----GACUAUAGUUUAUAAAAGUGACCAUCGUAUC------- .((..((((....((((((.....))))))..))))..))(((((.....(((((....))))).........((-----.(((............))).))....)))))------- ( -30.30) >consensus GAGUGGUGUCUAAGGUCAGGGACACUGACCGCACACUGCUGAUACCAGAGAGUUGGAAGCAAGUGCAAAUAAAGU_____AACUAUAGUUUAUGAAAGUGACCAUCGUAUC_______ ...(((((((...((((((.....))))))((.....)).)))))))....(.(((..((....)).......................((((....))))))).)............ (-26.43 = -26.43 + 0.00)


| Location | 19,962,505 – 19,962,603 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.08 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -23.89 |
| Energy contribution | -23.95 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19962505 98 - 20766785 UUUCC------ACCGAGCCACCGGGGGAUUGGUGUCUAAGGUCAGGGACACUGACCGCACACCGCUGAUACCAGACAGUUGGAAGCAAGUGCGAAAAAAGUACG .((((------.(((......)))))))..(((((....((((((.....))))))..)))))(((....((((....)))).)))..((((.......)))). ( -34.70) >DroSec_CAF1 3674 87 - 1 UUGCC------A--------CCGGGGGAGUGGUGUCUAAGGUCAGGGACACUGACCGCAAACUGCUGAUACCAGAGAGUUGGAAGCAGGUGCAAAUAAAGU--- ..(((------(--------((....).)))))......((((((.....))))))(((..(((((....((((....)))).))))).))).........--- ( -29.30) >DroSim_CAF1 3668 95 - 1 UUGCC------ACCGAGCCACCGGGGGAGUGGUGUCUAAGGUCAGGGACACUGACCGCACACUGCUGAUACCAGAGAGUUGGAAGCAACUGCAAAUAAAGU--- ((.((------.(((......))))).))(((((((...((((((.....))))))((.....)).)))))))...(((((....)))))...........--- ( -32.90) >DroYak_CAF1 3628 100 - 1 UUUCCACAGCCACAGAGCCACCGAGGGCUUGGUGUCUAAGGUCAGGGACACUGACCACACAGCGCCGAUACCAGCGAGUUGGAAGCAGCUGCGAAUAAAG---- .......((.(((.(((((......))))).))).))..((((((.....)))))).(.((((.(((((........))))).....)))).).......---- ( -32.40) >consensus UUGCC______ACCGAGCCACCGGGGGAGUGGUGUCUAAGGUCAGGGACACUGACCGCACACCGCUGAUACCAGAGAGUUGGAAGCAACUGCAAAUAAAGU___ ....................((((.....(((((((...((((((.....))))))((.....)).))))))).....))))..((....))............ (-23.89 = -23.95 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:33 2006