| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,949,089 – 19,949,204 |
| Length | 115 |
| Max. P | 0.821799 |

| Location | 19,949,089 – 19,949,204 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.57 |
| Mean single sequence MFE | -37.83 |
| Consensus MFE | -13.24 |
| Energy contribution | -14.10 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
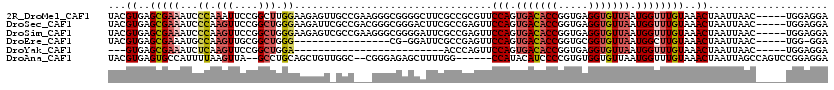
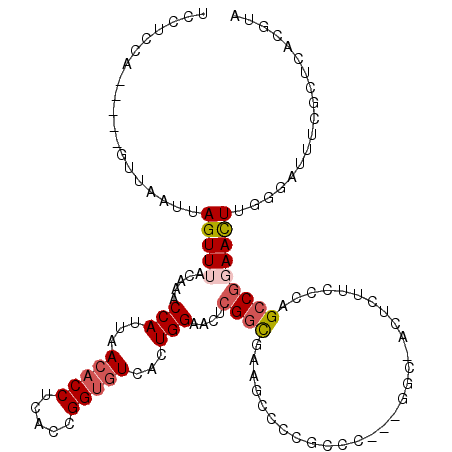
>2R_DroMel_CAF1 19949089 115 + 20766785 UACGUGAGCGAAAUCCCAAAUUCCGGCUUGGAAGAGUUGCCGAAGGGCGGGGCUUCGCCGCGUUCCAGUGACACCGGUGAGGUGUUAAUGGUUUGUAAACUAAUUAAC-----UGGAGGA .(((((.(((((..(((...((((.....)))).....(((....))))))..))))))))))((((((((((((.....))))))..(((((....)))))....))-----))))... ( -49.40) >DroSec_CAF1 32098 115 + 1 UACGUGAGCGAAAUCCCAAGUUCCGGCUGGGAAGAUUCGCCGACGGGCGGGACUUCGCCGAGUUCCAGUGACACCGGUGAGGUGUUAAUGGUUUGUAAACUAAUUAAC-----UGGAGGA ..(((..(((((.(((((.((....)))))))...)))))..)))(((((....)))))...(((((((((((((.....))))))..(((((....)))))....))-----))))).. ( -42.40) >DroSim_CAF1 34383 115 + 1 UACGUGAGCGAAAUCCCAAGUUCCGGCUGGGAAGAGUCGCCGAAGGGCGGGGAUUCGCCGAGUUCCAGUGACACCGGUGAGGUGUUAAUGGUUUGUAAACUAAUUAAC-----UGGAGGA ....((.(((((.((((...((((.....))))....((((....)))))))))))))))..(((((((((((((.....))))))..(((((....)))))....))-----))))).. ( -44.70) >DroEre_CAF1 31188 97 + 1 UACGUGAGCGAAAUGCCAAGUUGCGGCUGGG----------------CG-GGAUUCGCCGAGUUCCAGUGACACCGGUGCGGUGUUAAUGGCUUGUAAACUAAUUAAC-----UGG-GGA ...((..((((...((((..(((..(((.((----------------((-.....)))).)))..)))((((((((...)))))))).))))))))..))........-----...-... ( -30.80) >DroYak_CAF1 29887 87 + 1 ---GUGAGCGAAAUCUCAAGUUCCGGCUGGA-------------------------ACCCAGUUCCAGUGACACCGGUGAGGUGUUAAUGGUUUGUAAACUAAUUAAC-----UGGAGGA ---.((((......))))...(((.(((((.-------------------------..)))))((((((((((((.....))))))..(((((....)))))....))-----))))))) ( -27.30) >DroAna_CAF1 31408 110 + 1 UACGUGAGUGCCAUUUUAAGUUA--GCCUGCAGCUGUUGGC--CGGGAGAGCUUUUGG------CCAUACAUCCCCGUGUGGUGUUAAUGGUUUGUAAACUAAUUAGCCAGUCCGGAGGA ...(((.(.((((....(((((.--.((((..((.....))--))))..))))).)))------)).))).((((((..((((.....(((((....)))))....))))...))).))) ( -32.40) >consensus UACGUGAGCGAAAUCCCAAGUUCCGGCUGGGAAGAGU_GCC___GGGCGGGGAUUCGCCGAGUUCCAGUGACACCGGUGAGGUGUUAAUGGUUUGUAAACUAAUUAAC_____UGGAGGA ...((..(((((...((.(((....))).)).................................(((.(((((((.....))))))).))))))))..)).................... (-13.24 = -14.10 + 0.86)
| Location | 19,949,089 – 19,949,204 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.57 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -10.19 |
| Energy contribution | -11.00 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
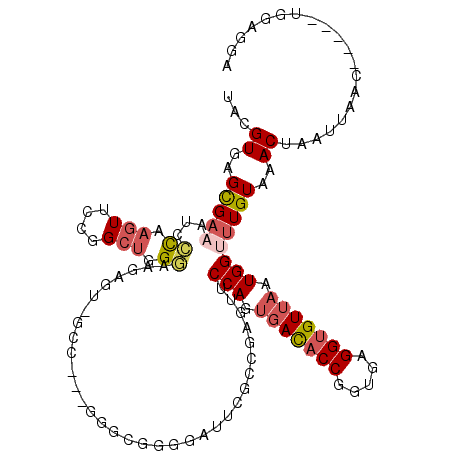
>2R_DroMel_CAF1 19949089 115 - 20766785 UCCUCCA-----GUUAAUUAGUUUACAAACCAUUAACACCUCACCGGUGUCACUGGAACGCGGCGAAGCCCCGCCCUUCGGCAACUCUUCCAAGCCGGAAUUUGGGAUUUCGCUCACGUA ...((((-----((......(((....))).....(((((.....))))).))))))(((.((((((..((((...((((((...........))))))...))))..))))).).))). ( -37.20) >DroSec_CAF1 32098 115 - 1 UCCUCCA-----GUUAAUUAGUUUACAAACCAUUAACACCUCACCGGUGUCACUGGAACUCGGCGAAGUCCCGCCCGUCGGCGAAUCUUCCCAGCCGGAACUUGGGAUUUCGCUCACGUA ...((((-----((......(((....))).....(((((.....))))).))))))....((((((((((((....(((((...........)))))....))))))))))))...... ( -39.20) >DroSim_CAF1 34383 115 - 1 UCCUCCA-----GUUAAUUAGUUUACAAACCAUUAACACCUCACCGGUGUCACUGGAACUCGGCGAAUCCCCGCCCUUCGGCGACUCUUCCCAGCCGGAACUUGGGAUUUCGCUCACGUA ...((((-----((......(((....))).....(((((.....))))).))))))....((((((..((((...((((((...........))))))...))))..))))))...... ( -34.60) >DroEre_CAF1 31188 97 - 1 UCC-CCA-----GUUAAUUAGUUUACAAGCCAUUAACACCGCACCGGUGUCACUGGAACUCGGCGAAUCC-CG----------------CCCAGCCGCAACUUGGCAUUUCGCUCACGUA ...-(((-----((......(((....))).....((((((...)))))).))))).....((((((...-.(----------------((.((......)).)))..))))))...... ( -23.10) >DroYak_CAF1 29887 87 - 1 UCCUCCA-----GUUAAUUAGUUUACAAACCAUUAACACCUCACCGGUGUCACUGGAACUGGGU-------------------------UCCAGCCGGAACUUGAGAUUUCGCUCAC--- ....(((-----(((.....(....)...(((.(.(((((.....))))).).)))))))))((-------------------------(((....))))).((((......)))).--- ( -24.00) >DroAna_CAF1 31408 110 - 1 UCCUCCGGACUGGCUAAUUAGUUUACAAACCAUUAACACCACACGGGGAUGUAUGG------CCAAAAGCUCUCCCG--GCCAACAGCUGCAGGC--UAACUUAAAAUGGCACUCACGUA (((.(((...(((.((((..(((....))).))))...)))..))))))(((.(((------((............)--)))))))((((.((((--((........)))).)))).)). ( -22.60) >consensus UCCUCCA_____GUUAAUUAGUUUACAAACCAUUAACACCUCACCGGUGUCACUGGAACUCGGCGAAGCCCCGCCC___GGC_ACUCUUCCCAGCCGGAACUUGGGAUUUCGCUCACGUA ...................(((((.....(((...(((((.....)))))...)))....((((.............................))))))))).................. (-10.19 = -11.00 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:20 2006