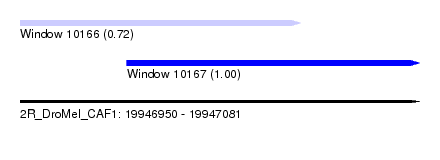
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,946,950 – 19,947,081 |
| Length | 131 |
| Max. P | 0.995172 |

| Location | 19,946,950 – 19,947,042 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.03 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -16.94 |
| Energy contribution | -16.80 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
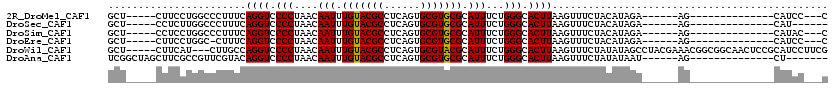
>2R_DroMel_CAF1 19946950 92 + 20766785 GCU-----CUUCCUGGCCCUUUCAGGUCCCCUAACAAUUUGUACGCCUCAGUGCGUGCGCAUUUCUGGGCACUUAAGUUUCUACAUAGA------AG--------------CAUCC---C (((-----(..(((((.....)))))..............((((((......))))))........))))......((((((....)))------))--------------)....---. ( -23.50) >DroSec_CAF1 30005 89 + 1 GCU-----CCUCUUGGCCCUUUCAGGUCCCCUAACAAUUUGUACGCCUCAGUGCGUGCGCAUUUCUGGGCACUUAAGUUUCUACAUAGA------AG--------------CAU------ (((-----......)))......((((.(((....(((.(((((((......))))))).)))...))).))))..((((((....)))------))--------------)..------ ( -23.00) >DroSim_CAF1 32265 92 + 1 GCU-----CCUCCUGGCCCUUUCAGGUCCCCUAACAAUUUGUACGCCUCAGUGCGUGCGCAUUUCUGGGCACUUAAGUUUCUACAUAGA------AG--------------CAUAC---C (((-----(..(((((.....)))))..............((((((......))))))........))))......((((((....)))------))--------------)....---. ( -23.10) >DroEre_CAF1 29064 91 + 1 GCU-----CUUCCUGGC-CUUUCAGGUCCCCUAACAAUUUGUACGCCUCAGUGCGUGCGCAUUUCUGGGCACUUAAGUUUCUACAUAGA------AG--------------CAUCC---C (((-----(..(((((.-...)))))..............((((((......))))))........))))......((((((....)))------))--------------)....---. ( -23.50) >DroWil_CAF1 28415 112 + 1 GCU-----CUUCAU---CUUGCCAGGUCCCCUAACAAUUUGUACGCCUCAGUGCGUACGCAUUUCUGGGCACUUAAGUUUCUAUAUAGCCUACGAAACGGCGGCAACUCCGCAUCCUUCG ...-----......---.(((((((((.(((....(((.(((((((......))))))).)))...))).)))).............(((........)))))))).............. ( -28.40) >DroAna_CAF1 28820 93 + 1 UCGGCUAGCUUCGCCGUUCGUACAGGUCCCCUAACAAUUUGUACGCCUCAGUGCGUGCGCAUUUCUGGGCACUUAAGUUUCUAUAUAAU------AG--------------CU------- .((((.......)))).......((((.(((....(((.(((((((......))))))).)))...))).))))...............------..--------------..------- ( -22.10) >consensus GCU_____CUUCCUGGCCCUUUCAGGUCCCCUAACAAUUUGUACGCCUCAGUGCGUGCGCAUUUCUGGGCACUUAAGUUUCUACAUAGA______AG______________CAUCC___C .......................((((.(((....(((.(((((((......))))))).)))...))).)))).............................................. (-16.94 = -16.80 + -0.14)

| Location | 19,946,985 – 19,947,081 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 73.89 |
| Mean single sequence MFE | -20.19 |
| Consensus MFE | -15.62 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 19946985 96 + 20766785 GUACGCCUCAGUGCGUGCGCAUUUCUGGGCACUUAAGUUUCUACAUAGAAGCAUCCCAUUCCCAUGCUCAUGCCCAUGCCCUCACCCUCACUCCAA ((((((......))))))(((....((((((.....((((((....))))))....(((....)))....)))))))))................. ( -24.20) >DroSec_CAF1 30040 78 + 1 GUACGCCUCAGUGCGUGCGCAUUUCUGGGCACUUAAGUUUCUACAUAGAAGCAU-----------------UCCCAUGCCCUCAUCCUGUC-CCAU ((((((......))))))(((.....(((((.....((((((....))))))..-----------------.....)))))......))).-.... ( -21.10) >DroSim_CAF1 32300 95 + 1 GUACGCCUCAGUGCGUGCGCAUUUCUGGGCACUUAAGUUUCUACAUAGAAGCAUACCAUUCCCAUGCCCAUGCCCAUGCCCUCAUCCUGUC-CCAU ((((((......))))))(((....((((((.....((((((....))))))....(((....)))....)))))))))............-.... ( -24.20) >DroEre_CAF1 29098 83 + 1 GUACGCCUCAGUGCGUGCGCAUUUCUGGGCACUUAAGUUUCUACAUAGAAGCAUCCCUUUCCCAUUCCCA------------CUCCCACUC-CCAU ((((((......)))))).......((((.......((((((....))))))........))))......------------.........-.... ( -17.46) >DroAna_CAF1 28860 83 + 1 GUACGCCUCAGUGCGUGCGCAUUUCUGGGCACUUAAGUUUCUAUAUAAUAGCU----AUAGCCAGUCCCCCGCCAACUUUUUCAACC--------- ((((((......))))))((....((((((......))..(((((.......)----))))))))......))..............--------- ( -14.00) >consensus GUACGCCUCAGUGCGUGCGCAUUUCUGGGCACUUAAGUUUCUACAUAGAAGCAU_CCAUUCCCAUGCCCA_GCCCAUGCCCUCACCCUCUC_CCAU ((((......))))((((.((....)).))))....((((((....))))))............................................ (-15.62 = -16.02 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:17 2006