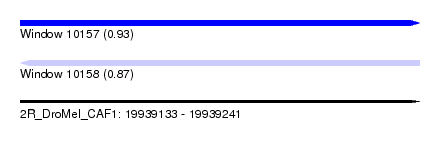
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,939,133 – 19,939,241 |
| Length | 108 |
| Max. P | 0.927189 |

| Location | 19,939,133 – 19,939,241 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.79 |
| Mean single sequence MFE | -41.72 |
| Consensus MFE | -28.65 |
| Energy contribution | -29.97 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
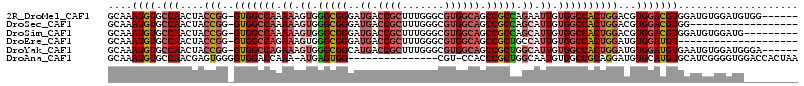
>2R_DroMel_CAF1 19939133 108 + 20766785 GCAAAUGUGCCAACUACCGG-GUGGCCAAAAAGUGGGCGGGAUGACCGCUUUGGGCGUGGCAGCCGCCAGAAUUGUGGCCACUGGACGUGGACGUGGAUGUGGAUGUGG------ (((.((((.(((.(((((.(-(((((((...(((.(((((..((.((((.......)))))).)))))...))).)))))))).)..))))...)))))))...)))..------ ( -42.10) >DroSec_CAF1 22293 96 + 1 GCAAAUGUGCCAACUACCGG-GUGGCCAAAAAGUGGGCGGGAUGACCGCUUUGGGCGUGGCAGCCGCCAGCAUUGUGGCCACUGGACGUGGACGUGG------------------ ....((((.(((....((((-.((((((.((.((.(((((..((.((((.......)))))).))))).)).)).))))))))))...)))))))..------------------ ( -41.30) >DroSim_CAF1 22606 105 + 1 GCAAAUGUGCCAACUACCGG-GUGGCCAAAAAGUGGGCGGGAUGACCGCUUUGGGCGUGGCAGCCGCCAGCAUUGUGGCCACUGGACGUGGACGUGGAUGUGGAUG--------- (((.((((.(((....((((-.((((((.((.((.(((((..((.((((.......)))))).))))).)).)).))))))))))...)))))))...))).....--------- ( -42.60) >DroEre_CAF1 21302 94 + 1 GCAAAUGUGCCAACUACCGG-GUGGCCAGAAAGUGGGCGGGAUGACCGCUUUGGGCGUGGCAGCCGCUGCCAUUGUGGCCACUGGAUGUGGAUGU-------------------- (((....)))((.(((((.(-(((((((.((...((((((.....))))))..(((((((...)))).))).)).)))))))).)..)))).)).-------------------- ( -39.00) >DroYak_CAF1 19814 108 + 1 GCAAAUGUGCCAACUACCGG-GUGGCCAGAAAGUGGGCGGCAUGACCGCUUUGGGCGUGGCAGCCGCUGGCAUUGUGGCCACUGGAUGUGGAUGUGAAUGUGGAUGGGA------ .........(((.(((((.(-(((((((.((.((.((((((.((.((((.......)))))))))))).)).)).)))))))).)..((........)))))).)))..------ ( -46.20) >DroAna_CAF1 20177 98 + 1 GCAAAUGUGCCAACGAGUGGGGUGGACCAAA-AUGAGUGG---------------CGU-CCACCCGCUGGCAAUGUGGCCGCAGGAUGUGCAUGUGCAUCGGGGUGGACCACUAA ........((((.....(((......)))..-.....)))---------------)((-((((((((.(((......)))))..((((((....)))))).))))))))...... ( -39.10) >consensus GCAAAUGUGCCAACUACCGG_GUGGCCAAAAAGUGGGCGGGAUGACCGCUUUGGGCGUGGCAGCCGCCAGCAUUGUGGCCACUGGACGUGGACGUGGAUGUGGAUG_________ ....((((.(((....(((..(((((((.((.((.(((((..((.((((.......)))))).))))).)).)).))))))))))...))))))).................... (-28.65 = -29.97 + 1.32)


| Location | 19,939,133 – 19,939,241 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.79 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -19.02 |
| Energy contribution | -20.90 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
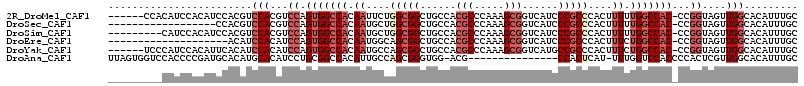
>2R_DroMel_CAF1 19939133 108 - 20766785 ------CCACAUCCACAUCCACGUCCACGUCCAGUGGCCACAAUUCUGGCGGCUGCCACGCCCAAAGCGGUCAUCCCGCCCACUUUUUGGCCAC-CCGGUAGUUGGCACAUUUGC ------................(((.((..((.(((((((.((....(((((.((.(.(((.....)))).))..)))))....)).)))))))-..))..)).)))........ ( -33.20) >DroSec_CAF1 22293 96 - 1 ------------------CCACGUCCACGUCCAGUGGCCACAAUGCUGGCGGCUGCCACGCCCAAAGCGGUCAUCCCGCCCACUUUUUGGCCAC-CCGGUAGUUGGCACAUUUGC ------------------....(((.((..((.(((((((.((.(..(((((.((.(.(((.....)))).))..)))))..).)).)))))))-..))..)).)))........ ( -33.20) >DroSim_CAF1 22606 105 - 1 ---------CAUCCACAUCCACGUCCACGUCCAGUGGCCACAAUGCUGGCGGCUGCCACGCCCAAAGCGGUCAUCCCGCCCACUUUUUGGCCAC-CCGGUAGUUGGCACAUUUGC ---------.............(((.((..((.(((((((.((.(..(((((.((.(.(((.....)))).))..)))))..).)).)))))))-..))..)).)))........ ( -33.20) >DroEre_CAF1 21302 94 - 1 --------------------ACAUCCACAUCCAGUGGCCACAAUGGCAGCGGCUGCCACGCCCAAAGCGGUCAUCCCGCCCACUUUCUGGCCAC-CCGGUAGUUGGCACAUUUGC --------------------....(((...((.(((((((...((((((...))))))........((((.....))))........)))))))-..))....)))......... ( -29.80) >DroYak_CAF1 19814 108 - 1 ------UCCCAUCCACAUUCACAUCCACAUCCAGUGGCCACAAUGCCAGCGGCUGCCACGCCCAAAGCGGUCAUGCCGCCCACUUUCUGGCCAC-CCGGUAGUUGGCACAUUUGC ------..................(((...((.(((((((.((.....(((((((.(.(((.....)))).)).))))).....)).)))))))-..))....)))......... ( -31.10) >DroAna_CAF1 20177 98 - 1 UUAGUGGUCCACCCCGAUGCACAUGCACAUCCUGCGGCCACAUUGCCAGCGGGUGG-ACG---------------CCACUCAU-UUUGGUCCACCCCACUCGUUGGCACAUUUGC ...((((((((......(((....))).....)).))))))..(((((((((((((-..(---------------(((.....-..)))).....)))))))))))))....... ( -37.90) >consensus _________CAUCCACAUCCACAUCCACAUCCAGUGGCCACAAUGCCAGCGGCUGCCACGCCCAAAGCGGUCAUCCCGCCCACUUUUUGGCCAC_CCGGUAGUUGGCACAUUUGC ........................(((...((.(((((((.((....(((((......(((.....)))......)))))....)).)))))))...))....)))......... (-19.02 = -20.90 + 1.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:07 2006