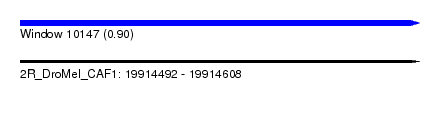
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 19,914,492 – 19,914,608 |
| Length | 116 |
| Max. P | 0.902910 |

| Location | 19,914,492 – 19,914,608 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 73.34 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -18.03 |
| Energy contribution | -19.17 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
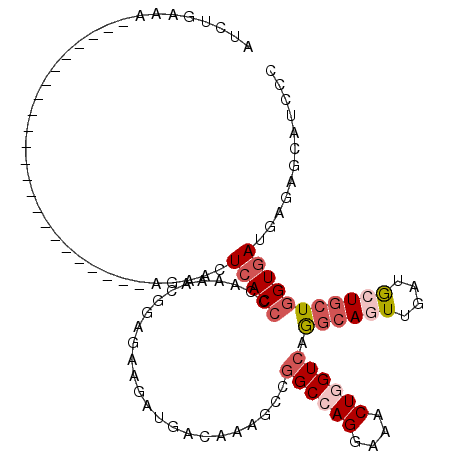
>2R_DroMel_CAF1 19914492 116 + 20766785 GUCUGAAAGACCCUUCGAAAAGAAGCUAUCGAGAACUCACCCCAAAACGGAGAAGAUGACAAAGCCGGCCAGGAAACUGGUCAAGCAGUUGAUGCUGCUGGUGAUGAGAGCAUCGC .((((...((..((((.....))))...))((....)).........))))...((((.((..(((((((((....)))))).((((((....)))))))))..))....)))).. ( -36.50) >DroSim_CAF1 5103 116 + 1 AUCUGAAAGACCCUUUGAAGAGAAGCUAUUGAGAACUCACCCCAAAACGGAGAAGAUGACAAAGCCGGCCAGGAAACUGGUCAGGCAGUUGAUGCUGCUGGUGAUGAGGGCAUCGC .((((((((...))))...(((...((....))..))).........))))...((((.((..(((((((((....)))))).((((((....)))))))))..))....)))).. ( -34.40) >DroEre_CAF1 3788 92 + 1 AUGUGAAA------------------------GAACUCACCCCAAAACGGAGAAGAUGACAAAGCCGGCCAGGAAGCUGGUCAGGCAGUUGAUGCUGCUGGUGAUGAGGGCAUCCC ..((((..------------------------....)))).((.....))....((((.((..(((((((((....)))))).((((((....)))))))))..))....)))).. ( -29.60) >DroWil_CAF1 12099 92 + 1 AUCUCAG------------------------UGUACUCACCCCAAUACUGAGAAGAUGACAAAUCCGGCCAGGAAACUCGUCAAACAAUUAAUACUACUCGUUAUAAGAGCAUCUC .((((((------------------------(((..........)))))))))(((((........(((.((....)).)))...............(((.......)))))))). ( -23.10) >DroYak_CAF1 3732 92 + 1 AUCUGAAG------------------------AGACUCACCCCAAGACGGAGAAGAUGACAAAGCCGGCGAGGAAGCUGGUCAGGCAGUUGAUGCUGCUGGUGAUGAGGGCAUCCC ........------------------------....(((((((.....)).............((((((......))))))..((((((....)))))))))))...(((...))) ( -25.00) >DroAna_CAF1 3722 94 + 1 U--------------UGAAAGG--------AAGAACUCACCCCAAAACUGAAAAUAUUACAAACCCAGCCAGAAAACUGGUCAGGCAAUUGAUGCUGCUGGUGAUUAGAGCAUCUC .--------------.....((--------...........))....................((..(((((....)))))..)).....((((((.(((.....))))))))).. ( -16.70) >consensus AUCUGAAA_______________________AGAACUCACCCCAAAACGGAGAAGAUGACAAAGCCGGCCAGGAAACUGGUCAGGCAGUUGAUGCUGCUGGUGAUGAGAGCAUCCC ....................................(((((.........................((((((....)))))).((((((....)))))))))))............ (-18.03 = -19.17 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:56 2006